Drug Metabolism Analysis Services
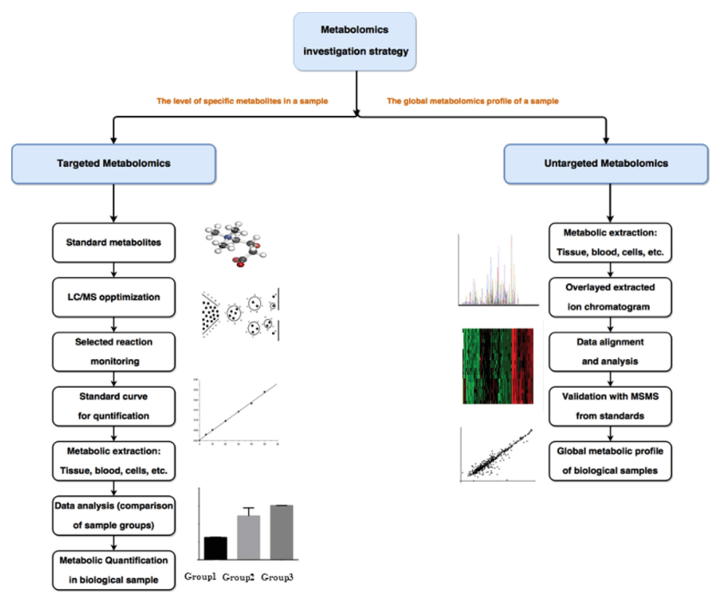
In drug discovery, reliable identification of the chemical structure of metabolites is essential for drug compound evaluation. In the drug discovery phase, combining high-resolution accurate mass data with MS/MS fragmentation patterns for generating molecular formulae facilitates the understanding of inferred metabolite structures. The use of metabolite structure prediction software in combination facilitates the structural identification of metabolites.
Creative Proteomics offers metabolomic solutions for drug metabolite analysis. We can purify and structurally characterize metabolites from complex biological samples.
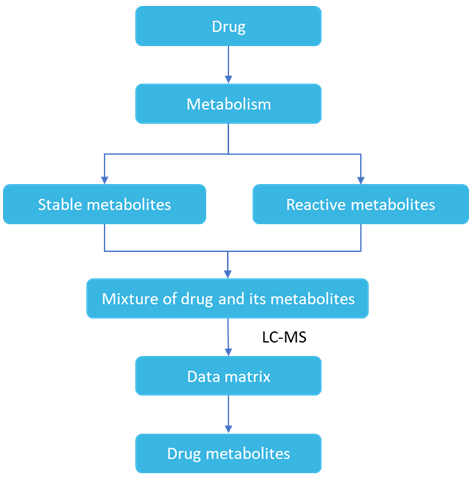
The processes of absorption, distribution, metabolism and excretion of drugs in living organisms. Most drugs are metabolized mainly by the liver, and the metabolites are excreted by the kidneys in the urine.
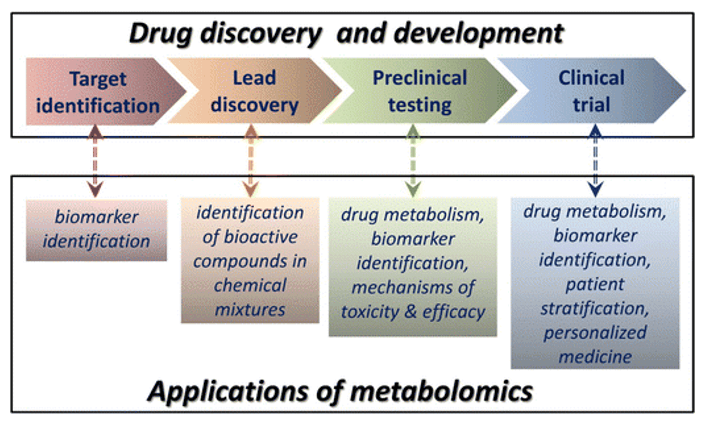
In preclinical or clinical drug metabolism studies, the overall characteristics of drug metabolism in vivo as reflected by whole animal and human studies can provide useful information on drug metabolism in humans. The search for active metabolites of drugs in vivo is an important tool to elucidate the material basis of drug efficacy. The search for phase II metabolic conjugates can provide a theoretical basis for the study of drug excretion and for the relief of drug intoxication. Therefore, the analysis of drug metabolites has positive significance in elucidating the pharmacological mechanism of drugs, clarifying the material basis of drug activity, and guiding the rational use of drugs in clinical practice.
Drug Metabolism Analysis Technology Platform
LC-MS /MS uses multi-stage mass spectrometry data tandem, with common scans such as daughter ion scan, parent ion scan, neutral loss scan, multiple reaction monitoring (MRM) or selected reaction monitoring (SRM).
| Product ion scan | The cleavage pattern of the drug backbone is first obtained by obtaining the daughter ion spectrum of the parent drug, and then the daughter ion spectrum of the metabolite is obtained and linked to the biotransformation pattern and possible metabolic sites to obtain the possible structure of the metabolite. It is suitable for analyzing the structural information of metabolites. |
| Daughter ion scan | Tracing the origin of the daughter ion fragments allows screening of a class of compounds that produce the same type of fragment ions. |
| Neutral loss scan | Determines the important functional groups contained in the original molecule by detecting the exact mass of the missing fragment. |
| MRM or SRM | Analyze complex biological samples for a specific mass number of analytes to be measured, allowing quantitative analysis of trace metabolic components at low concentrations. |
| Information depended acquire (IDA) | Full scan, neutral loss scan, multiple reaction monitoring, etc. are performed by pre-set conditions of ion intensity, mass number and retention time, and eligible fragment ions are selected for multi-stage mass spectrometry to obtain specific structural information. |
The blank control method is the most commonly used method for metabolite studies. A blank control group and a drug administration group are set up simultaneously, and a peak present in the drug administration group but not in the blank control group is screened, then the compound represented by that peak is a possible metabolite. Drug metabolism usually adds or eliminates functional groups to the prototype drug, so the metabolites usually retain the backbone structure of the prototype drug. It is often assumed that the cleavage patterns of the two are similar and that similar cleavage reactions can occur. Some of the same neutral fragments are lost or identical fragment ions are formed during the mass spectrometry scan. Possible metabolites can be found by using techniques such as parent ion scans, neutral loss scans, combined with analysis of MSn spectra of drugs and metabolites to find identical characteristic information.
- Drug metabolites identification
Partial structures are deduced from the similarity of the cleavage pattern of the drug metabolites with the prototype drug and their specific cleavage patterns, and specific reaction sites are determined by comparison with the prototype drug. The type of biotransformation can be determined by the difference in mass number between the metabolite and the prototype drug.
If you would like to learn more, please contact us and we look forward to working with you.
For Research Use Only. Not for use in diagnostic procedures.