What is Polyamine?
Polyamines, including putrescine, spermidine, and spermine, are positively charged small molecules found in virtually all living cells. They are essential regulators of gene expression, protein synthesis, cell proliferation, apoptosis, and intracellular signaling. Because their metabolism is tightly linked to amino acid, methionine, and urea cycles, polyamines serve as key indicators of cellular metabolic states.
In research, subtle changes in polyamine concentration or turnover can reveal shifts in metabolic pathways, responses to environmental stress, or biological impacts of experimental interventions. This makes them valuable targets for studies in molecular biology, biochemistry, systems biology, and translational research.
Accurate polyamine profiling allows researchers to:
- Map biosynthetic and catabolic pathway activities.
- Identify metabolic signatures associated with specific biological states.
- Evaluate the impact of genetic, nutritional, or environmental factors on cellular metabolism.
Advantages of Polyamine Analysis Service
- Quantitative Accuracy: Our isotope-labeled tracing methods provide quantitation with CVs <10% across biological replicates, enabling precise flux measurements.
- High Throughput Capability: Analyze up to 200 samples per run with batch processing to meet high-volume project needs.
- Wide Dynamic Range: Detection range of 10 nM to 100 µM, ensuring both trace and abundant metabolites are captured effectively.
- Pathway-Integrated Interpretation: Integrated metabolic modeling and pathway visualization support downstream interpretation and hypothesis generation.
- Time-Resolved Kinetics: Kinetic analysis with time-course sampling from 5 min to 24 h post-substrate administration.
- Cross-Species Compatibility: Validated in human, mouse, rat, and other model organisms for translational research.
Polyamine Analysis Service Offered by Creative Proteomics
- Quantification of Core Polyamines: Accurate measurement of putrescine, spermidine, spermine, and cadaverine using isotope-labeled internal standards and LC-MS/MS.
- Metabolite Profiling in Polyamine Pathways: Detection of key precursors and derivatives such as ornithine, arginine, agmatine, N-acetylspermidine, and N-acetylspermine.
- Analysis of Catabolic and Enzyme-Associated Metabolites: Monitoring of degradation byproducts and intermediates related to enzymes like ODC, SAM decarboxylase, and PAO.
- Pathway-Integrated Metabolomics: Expanded profiling of arginine, methionine, and urea cycle metabolites including SAM, SAH, and MTA.
- Comparative Studies and Multi-Condition Analysis:Tailored data generation for treated vs. control samples, time-course studies, and environmental factor assessments.
- Custom Polyamine Panels: Flexible method development for rare or novel polyamines in species-specific or experimental research.
- Comprehensive Data Reporting: Deliverables include full quantitation, pathway enrichment analysis, and visual outputs such as heatmaps and PCA.
List of Detected Polyamine and Related Metabolites
| Category |
Representative Metabolites |
Notes / Pathways |
| Core Polyamines |
Putrescine, Spermidine, Spermine, Cadaverine |
Basic bioactive polyamines |
| Acetylated Polyamines |
N1-Acetylputrescine, N1-Acetylspermidine, N1-Acetylspermine, N8-Acetylspermidine, N8-Acetylspermine |
Catabolism intermediates |
| Methylated Polyamines |
N-Methylputrescine, N-Methylspermidine, Trimethylspermidine, N,N′-Dimethylspermidine |
Involved in methylation regulation |
| Polyamine Precursors |
Ornithine, Arginine, Lysine |
Enter biosynthesis via ODC, ADC, LDC |
| Decarboxylation Products |
Agmatine, 1,3-Diaminopropane, 1,5-Diaminopentane (Cadaverine), γ-Aminobutyric acid (GABA), β-Alanine |
Decarboxylation byproducts from amino acids |
| Enzyme-Linked Metabolites |
Decarboxylated SAM (dcSAM), SAM, SAH, MTA (5′-Methylthioadenosine) |
Linked to AdoMetDC and polyamine synthesis enzymes |
| Methionine Cycle Metabolites |
Methionine, Homocysteine, Cystathionine |
SAM donors and methylation metabolism |
| Urea Cycle Intermediates |
Citrulline, Ornithine, Argininosuccinate, Urea, Carbamoyl phosphate |
Nitrogen metabolism; precursor cycling |
| Stress- and Degradation-Related |
Creatinine, Creatine, Taurine, Hydroxyproline |
Indirect markers; linked to stress, turnover |
| Polyamine Oxidation Byproducts |
Hydrogen peroxide (H₂O₂)*, Aminoaldehydes (e.g., 3-aminopropanal)* |
Polyamine oxidase (PAO) pathway – optional detection |
| Microbial / Plant-Specific Polyamines |
Nor-spermidine, Thermospermine, Sym-norspermidine |
Found in specific microbial and plant systems |
| Cross-Pathway Biogenic Amines |
Tryptamine*, Serotonin* |
Intersection with tryptophan and neuromodulator metabolism (optional) |
*Note: Some compounds (e.g., reactive aldehydes, H₂O₂, serotonin) may require additional derivatization or specific detection modules and are available upon request.
Workflow for Polyamine Analysis Service
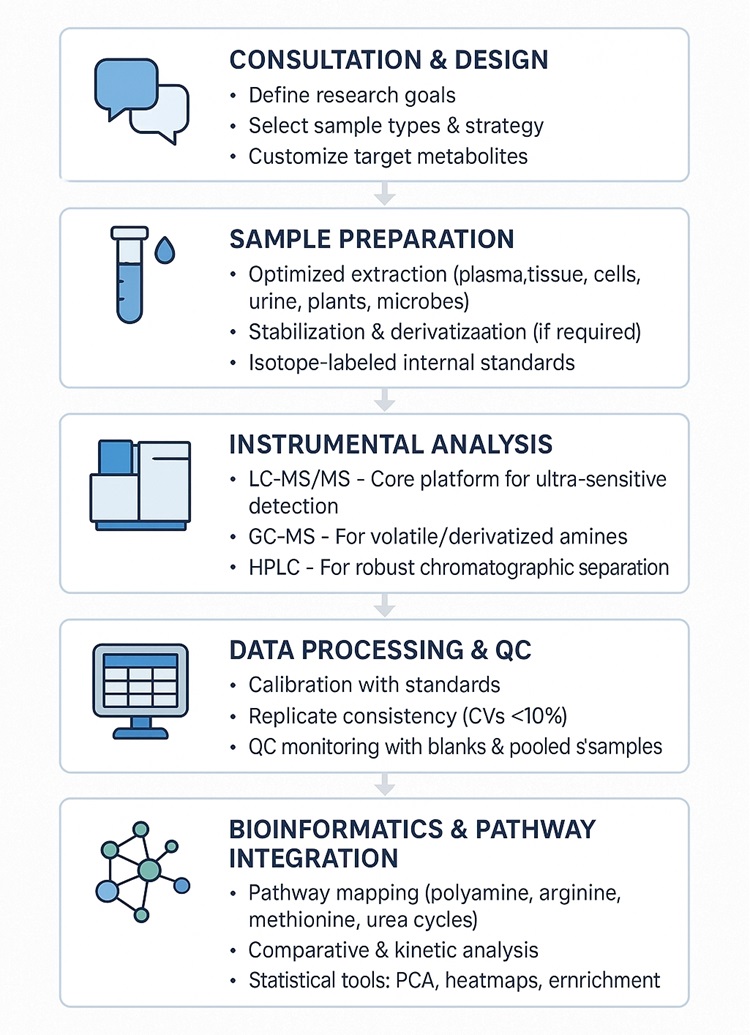
Technology Platform for Polyamine Analysis Service
LC-MS/MS – Our primary technique using SCIEX QTRAP® 6500+ and Thermo TSQ Altis™ for ultra-sensitive, targeted analysis of polyamines and their derivatives.
GC-MS – Ideal for volatile or derivatized amines, performed on Agilent 7890B-5977A systems with high resolution and reproducibility.
HPLC – Quantification after derivatization using the Agilent 1260 Infinity II HPLC system, ensuring reliable separation and detection.
Sample Requirements for Polyamine Analysis Service
| Sample Type |
Required Sample Amount |
Notes |
| Plasma / Serum |
100 – 200 µL |
Fresh or frozen, avoid repeated freeze-thaw cycles |
| Tissue (animal/plant) |
20 – 50 mg (wet weight) |
Snap-frozen preferred, stored at -80°C |
| Cell Pellets |
1 – 5 million cells |
Washed, pelleted, and frozen promptly |
| Urine |
1 – 2 mL |
Stored at -80°C, avoid contamination |
| Microbial Cultures |
10 – 50 mL culture volume or pellet equivalent |
Harvested at exponential phase recommended |
| Plant Extracts / Sap |
0.5 – 1 mL |
Clarified and frozen, filtered if necessary |
Applications of Polyamine Assay Service
Polyamine metabolism impacts T cell dysfunction in the oral mucosa of people living with HIV
Journal: Nature Communications
Published: 2023
Background
HIV infection causes persistent immune dysfunction and inflammation, especially in the oral mucosa, leading to increased oral diseases in people living with HIV. Metabolic pathways like polyamine synthesis influence T cell function and subset balance. This study investigates how altered polyamine metabolism during HIV infection disrupts T cell regulation, contributing to chronic inflammation and immune imbalance.
Methods
Our client leveraged Creative Proteomics's LC-MS/MS platforms to address critical challenges in HIV-associated metabolic and immune dysregulation research:
Comprehensive Salivary Metabolomics:
- Problem: Identifying HIV-driven metabolic perturbations in saliva.
- Solution: Creative Proteomics performed LC-MS-based metabolomic profiling (Ultimate 3000LC + Q Exactive MS, ESI ± modes) on methanol-extracted saliva, enabling the discovery of elevated polyamines (e.g., putrescine) and dysregulated arginine metabolism in HIV+ patients.
Targeted Polyamine Quantification:
- Problem: Validating HIV-induced polyamine biosynthesis in immune cells.
- Solution: Our UPLC-MRM/MS platform (Agilent 1290 UHPLC + 6495B QQQ) with dansyl chloride derivatization enabled precise quantification of intracellular putrescine, spermidine, and spermine, confirming ODC-1-dependent polyamine accumulation in HIV-infected CD4+ T cells.
Integration with Multi-Omics Data:
- Problem: Linking metabolic changes to transcriptional dysregulation.
- Solution: Creative Proteomics' data analysis pipeline (Metaboanalyst, Cytoscape) integrated LC-MS metabolomics with client-generated RNA-seq data, revealing correlations between ODC-1/EIF5A upregulation and polyamine overproduction.
Impact: By deploying our end-to-end metabolomics services (sample preparation → LC-MS analysis → bioinformatics), the client successfully:
- Identified HIV-specific salivary polyamine signatures.
- Validated ODC-1 as the driver of pathogenic polyamine synthesis.
- Demonstrated the therapeutic potential of polyamine pathway inhibition.
Results
Salivary Metabolome and Transcriptome Aberrations in HIV+ Individuals Metabolomic analysis revealed significantly elevated polyamine levels (e.g., putrescine) and dysregulated arginine metabolism in the saliva of HIV+ patients. Oral mucosal transcriptomics showed upregulated expression of ODC-1 (ornithine decarboxylase-1) and EIF5A (eukaryotic translation initiation factor 5A), with flow cytometry confirming increased ODC-1 and EIF5A protein levels in CD4+ T cells.
HIV Infection Disrupts TregDys/Th17 Balance In ex vivo HIV-infected tonsil organoid cultures (HTOC), Th17 cells (IL-17A+ ROR-γt+) were markedly reduced, while PD-1hi IFN-γ+ FOXP3+ TregDys expanded persistently, even after antiretroviral therapy (ART)
HIV Upregulates ODC-1, EIF5A, and Hypusination to Promote Polyamine Synthesis HIV infection increased ODC-1 protein, EIF5A, and its hypusination (a post-translational modification) in CD4+ T cells, accompanied by elevated intracellular polyamines (putrescine, spermidine, spermine).
Caspase-1 and IL-1β Mediate ODC-1 Upregulation HIV activated caspase-1 to induce IL-1β release, which upregulated ODC-1 via IL-1 receptor signaling in CD4+ T cells. Caspase-1 inhibition (VX-765) or IL-1β blockade reversed ODC-1 elevation and TregDys expansion.
ODC-1 Activity Drives TregDys Expansion ODC-1 inhibition (DFMO or shRNA) reduced polyamines, EIF5A expression, and TregDys populations but failed to restore Th17 loss, suggesting Th17 depletion is mediated by HIV-induced pyroptosis.
HIV-Induced Polyamine Biosynthesis Depends on ODC-1 LC-MS quantification confirmed significant increases in putrescine, spermidine, and spermine post-HIV infection, with ODC-1 inhibitors restoring normal polyamine levels.
Exogenous Polyamines Directly Promote TregDys Proliferation Supplementation with putrescine or spermidine upregulated TregDys proportions and AREG (amphiregulin) expression, enhancing proliferation (KI-67+). Conversely, blocking EIF5A hypusination (GC7) suppressed TregDys expansion.
Clinical Correlation Between TregDys/Th17 Ratio and Polyamine Levels HIV+ patients exhibited elevated TregDys/Th17 ratios in oral mucosa, positively correlating with salivary putrescine levels and CD4+ T cell hyperactivation (CD38+ HLA-DR+).
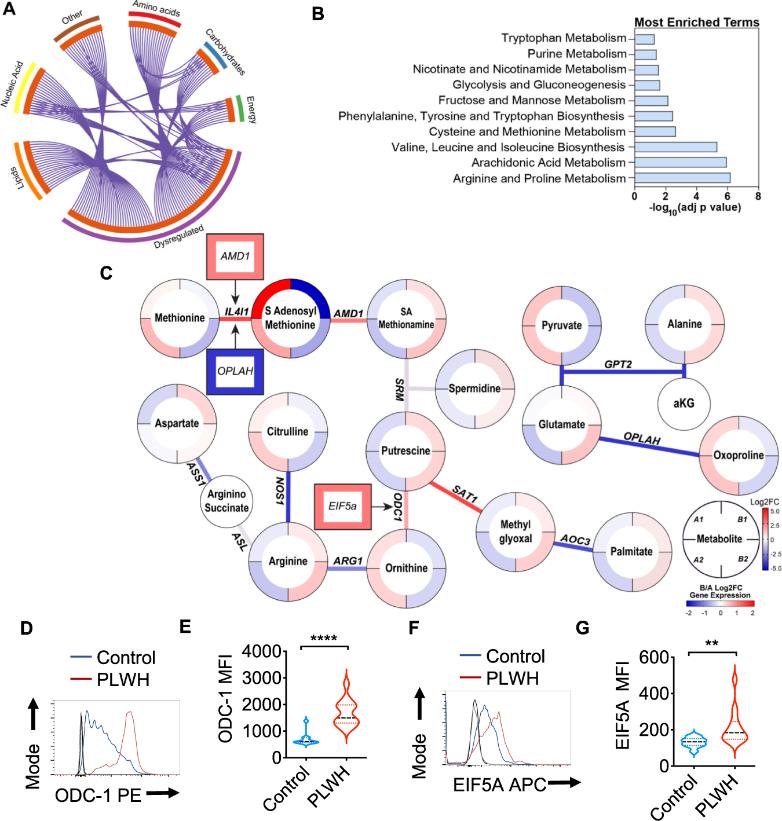 Metabolite levels were determined by LC-MS in saliva samples from healthy controls (Group A) and PLWH (Group B) (uninfected controls n = 26; PLWH; n = 40).
Metabolite levels were determined by LC-MS in saliva samples from healthy controls (Group A) and PLWH (Group B) (uninfected controls n = 26; PLWH; n = 40).
Conclusions
HIV infection drives oral mucosal immune dysregulation by activating the caspase-1/IL-1β axis to upregulate ODC-1, reprogramming polyamine metabolism to expand TregDys while depleting Th17 cells. Targeting polyamine pathways may represent a therapeutic strategy to mitigate HIV-associated chronic inflammation
Reference
- Mahalingam, S. S., et al. "Polyamine metabolism impacts T cell dysfunction in the oral mucosa of people living with HIV." Nature Communications 14.1 (2023): 399.
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal: Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.3390/antiox9111098
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z







 Metabolite levels were determined by LC-MS in saliva samples from healthy controls (Group A) and PLWH (Group B) (uninfected controls n = 26; PLWH; n = 40).
Metabolite levels were determined by LC-MS in saliva samples from healthy controls (Group A) and PLWH (Group B) (uninfected controls n = 26; PLWH; n = 40).