What Is Central Carbon Metabolism and Why Profile It?
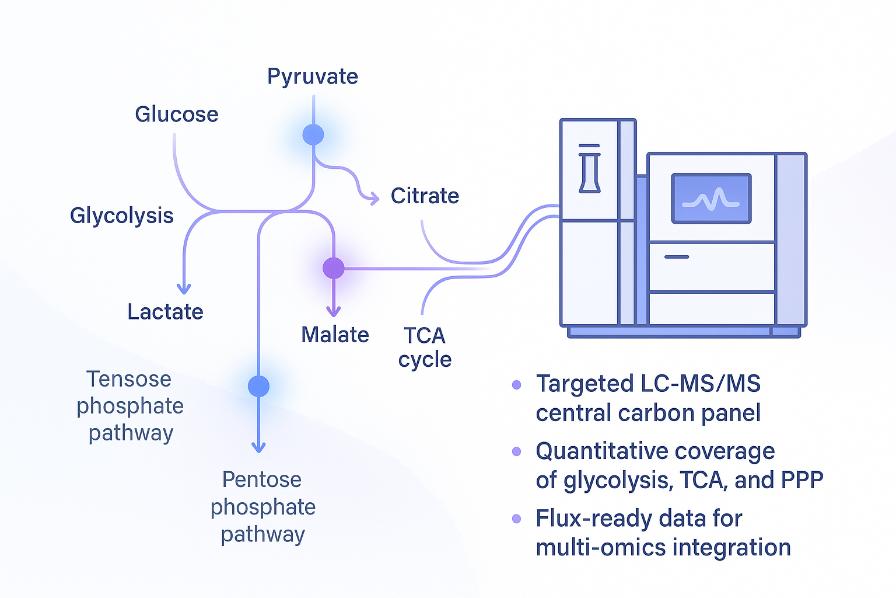
Central carbon metabolism is the cellular hub that converts nutrients into energy, redox cofactors, and building blocks for macromolecules, including glycan precursors. Glycolysis, the TCA cycle, and the pentose phosphate pathway work together to control ATP supply, redox balance, and carbon skeletons that support growth and product quality.
Because these pathways respond rapidly to media, genetic changes, and process stress, shifts in central carbon flux often appear before titer, viability, or glycan profiles change. Quantitative central carbon metabolism analysis therefore provides an early, pathway-level view of how a system is adapting and where optimization is possible. Typical questions this profiling can address include:
- How do key pathways respond to nutrient shifts, engineering, or small-molecule treatment?
- Does a given cell line or strain favor glycolytic or mitochondrial routes under specific conditions?
- Which steps in central carbon metabolism constrain productivity or robustness in fermentation or culture?
- How does carbon routing affect pools of glycan-related precursors and other biosynthetic intermediates?
Our Central Carbon Metabolism Quantification and Profiling
Creative Proteomics offers a modular central carbon metabolism analysis platform that can be tailored to your experimental system. Typical service modules include:
- Targeted central carbon metabolite quantification: Absolute or relative quantitation of key intermediates in glycolysis, TCA cycle, pentose phosphate pathway, anaplerotic routes, and related shunts.
- Stable isotope–based pathway flux analysis: 13C-labeled substrates (for example, glucose, glutamine, acetate) combined with time-course sampling and model-based flux estimation.
- Glycan precursor and cofactor profiling: Quantification of UDP-sugars, sugar phosphates, and redox cofactors relevant to glycan biosynthesis and cellular energy balance.
- Nicotinate and nicotinamide metabolite analysis: Targeted profiling of nicotinic acid, nicotinamide, NAD⁺, NADH, NADP⁺, NADPH, and related intermediates.
- Extracellular metabolite and nutrient balance: Measurement of substrate uptake and byproduct secretion to support carbon and redox balance calculations.
- Custom method development: Extension of analyte panels or isotope labeling strategies to fit unique pathways, engineered routes, or non-standard substrates.
Each module can be combined into an end-to-end study, from cell culture sampling to integrated pathway interpretation.
Analyte Coverage: Central Carbon Metabolites and Related Compounds
| Pathway / Class |
Representative analytes |
Notes / typical use |
| Glycolysis and related intermediates |
Glucose, glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, DHAP, glyceraldehyde-3-phosphate, 3-phosphoglycerate, 2-phosphoglycerate, phosphoenolpyruvate, pyruvate, lactate |
Core carbohydrate catabolism, energy generation, lactate formation, carbon entry into TCA. |
| TCA cycle and anaplerotic nodes |
Citrate, cis-aconitate, isocitrate, α-ketoglutarate, succinate, fumarate, malate, oxaloacetate, aspartate, glutamate, glutamine |
Mitochondrial function, anaplerosis, connection to amino acid biosynthesis and redox balance. |
| Pentose phosphate pathway and nucleotide precursors |
6-phosphogluconate, ribulose-5-phosphate, ribose-5-phosphate, xylulose-5-phosphate, sedoheptulose-7-phosphate, erythrose-4-phosphate |
NADPH generation, ribose supply for nucleotides, oxidative and non-oxidative PPP activity. |
| Glycan precursor–related metabolites |
Glucosamine-6-phosphate, N-acetylglucosamine-6-phosphate, UDP-glucose, UDP-galactose, UDP-N-acetylglucosamine, GDP-mannose |
Links central carbon metabolism to sugar nucleotides and glycan precursor pools. |
| Nicotinate and nicotinamide pathway metabolites |
Nicotinic acid, nicotinamide, nicotinamide mononucleotide (NMN), nicotinamide riboside (NR), NAD⁺, NADH, NADP⁺, NADPH |
Redox state, cofactor balance, and nicotinate/nicotinamide pathway activity. |
| Other supporting metabolites |
Acetyl-CoA (and selected acyl-CoAs where applicable), key amino acids, organic acids |
Integration of carbon and nitrogen balance, links to lipid synthesis and overflow metabolism. |
This panel can be expanded with additional targets on request, for example pathway-specific intermediates or custom sugar nucleotides.
Advantages of Central Carbon Metabolism Quantitative Analysis
- High-precision calibration – Targeted LC–MS/MS with isotope-labeled internal standards; calibration curves typically reach R² ≥ 0.99.
- Robust quality control – Pooled QC samples and internal standards keep intra-batch CVs commonly ≤ 15%, supporting confident group comparisons.
- Broad dynamic range – Optimized MRM methods cover 3–4 orders of magnitude, capturing both low-abundance intermediates and high-abundance end products in one run.
- Pathway-focused coverage – A curated panel spanning glycolysis, the TCA cycle, and the pentose phosphate pathway provides contiguous central carbon readouts.
- Flux-ready methods – The same setup supports steady-state quantitation and 13C isotopologue profiling, enabling straightforward extension to flux analysis.
- Integration-ready outputs – Data are delivered with stable identifiers and pathway annotations, ready for direct import into R, Python, and pathway analysis tools
Analytical Platform for Central Carbon Metabolism LC–MS/MS
Central carbon metabolites are quantified on a targeted LC–MS/MS platform built around UPLC systems and triple quadrupole mass spectrometers. The setup is optimized for polar metabolites in glycolysis, the TCA cycle, and the pentose phosphate pathway, with robust retention, sensitivity, and throughput for routine project work.
Instrumentation Overview
| Component |
Typical Configuration |
Purpose |
| UPLC system |
Waters ACQUITY UPLC I-Class / H-Class; Agilent 1290 II |
Precise gradients, stable retention of polar metabolites |
| MS detector |
SCIEX QTRAP 6500+; Agilent 6495C |
High-sensitivity MRM for targeted metabolomics |
| Ionization mode |
ESI, positive and negative with fast polarity switching |
Coverage of organic acids, sugar phosphates, amino acids |
Typical LC–MS/MS Settings for Central Carbon Metabolites
| Parameter |
Typical Setting |
Notes |
| Column |
2.1 × 100 mm, 1.7 µm HILIC (amide) |
Polar metabolites and sugar phosphates |
| Column temperature |
30–40 °C |
Stable retention and peak shape |
| Flow rate |
0.2–0.4 mL/min |
Adjusted to column and matrix |
| Injection volume |
2–10 µL |
Tuned to concentration and matrix complexity |
| Gradient length |
~10–20 min per sample |
Balance separation and throughput |
| Acquisition mode |
Scheduled MRM |
Sufficient data points per peak for dense panels |
| Internal standards |
Stable isotope–labeled analogs |
Correct matrix effects and normalize instrument drift |
| Calibration |
Multi-level external calibration |
Define linear range and LOQ for key metabolites |
| Batch QC |
Pooled QC, blanks, calibration set |
Monitor retention time, signal stability, and CV |
Targeted methods with isotope-labeled internal standards, multi-level calibration, and batch-level QC ensure that central carbon readouts are quantitative, reproducible, and suitable for downstream modeling and integration with other omics data.
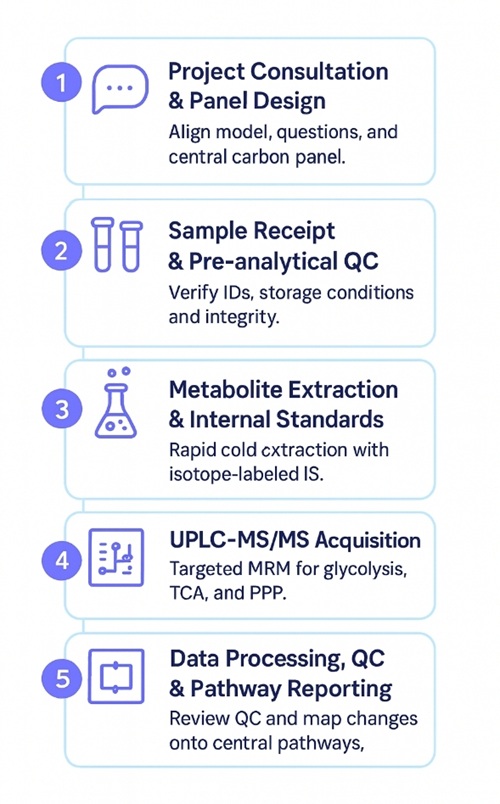
Workflow for Central Carbon Metabolism LC–MS/MS Panel
Sample Requirements for Central Carbon Panel Analysis
| Sample type |
Recommended amount |
Container |
Storage & transport notes |
| Blood / plasma / serum |
≥ 500 µL per sample |
Pre-labeled cryovial, anticoagulant as appropriate |
Keep frozen; ship on dry ice; avoid repeated freeze–thaw cycles. |
| Urine |
≥ 1 mL per sample |
Screw-cap tube or cryovial |
Mix well before aliquoting; freeze promptly; send on dry ice. |
| Solid tissue |
~ 200 mg per sample |
Pre-cooled tube |
Snap-freeze after collection; keep at ultra-low temperature; ship on dry ice. |
| Cultured cells |
≥ 1 × 10⁷ cells per sample |
Tube compatible with rapid quenching |
Quench metabolism rapidly; store pellets frozen; transport on dry ice. |
| Feces or digesta |
~ 500 mg per sample |
Leak-proof tube |
Freeze as soon as possible; ship on dry ice to preserve metabolite stability. |
For other matrices such as culture media, formulated products, or specialized biological fluids, tailored requirements can be defined during project setup.
Deliverables: What You Receive from Central Carbon Metabolism Analysis
Technical summary – Study design overview, sample list, and analytical methods used.
Raw data files – Original LC–MS/GC–MS instrument files for all samples and QC runs.
Processed data tables – Curated tables of metabolite intensities or concentrations, with sample IDs and normalization details.
Isotope tracing outputs (if applicable) – Isotopologue distributions and label fractions for tracer-based experiments.
Quality control report – Internal standard performance, calibration results, and batch QC metrics.
Pathway-level data views – Pathway-organized result tables or figures summarizing changes in central carbon and related metabolites.
Research Applications of Central Carbon Metabolism Profiling
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
Macrophage-associated lipin-1 promotes β-oxidation in response to proresolving stimuli
Schilke, R. M., Blackburn, C. M., Rao, S., Krzywanski, D. M., Finck, B. N., & Woolard, M. D.
Journal: Immunohorizons
Year: 2020
Lipin-1 regulates lipid catabolism in pro-resolving macrophages
Schilke, R. M., Blackburn, C. M., Rao, S., Krzywanski, D., Finck, B. N., & Woolard, M. D.
Journal: bioRxiv
Year: 2020
YAP mediates compensatory cardiac hypertrophy through aerobic glycolysis in response to pressure overload
Kashihara, T., Mukai, R., Oka, S. I., Zhai, P., Nakada, Y., Yang, Z., ... & Sadoshima, J.
Journal: The Journal of Clinical Investigation
Year: 2022
Inflammation primes the kidney for recovery by activating AZIN1 A-to-I editing
Heruye, S., Myslinski, J., Zeng, C., Zollman, A., Makino, S., Nanamatsu, A., ... & Hato, T.
Journal: bioRxiv
Year: 2023