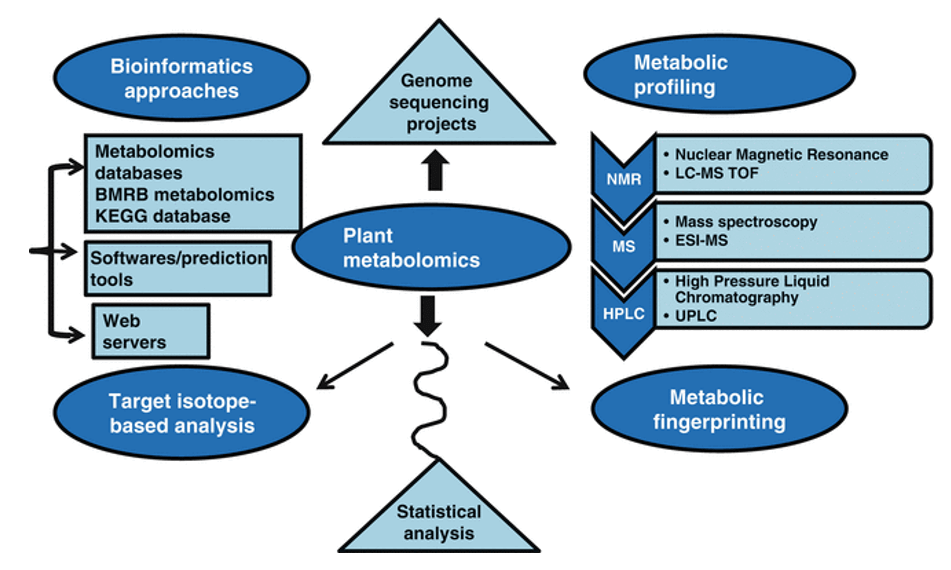
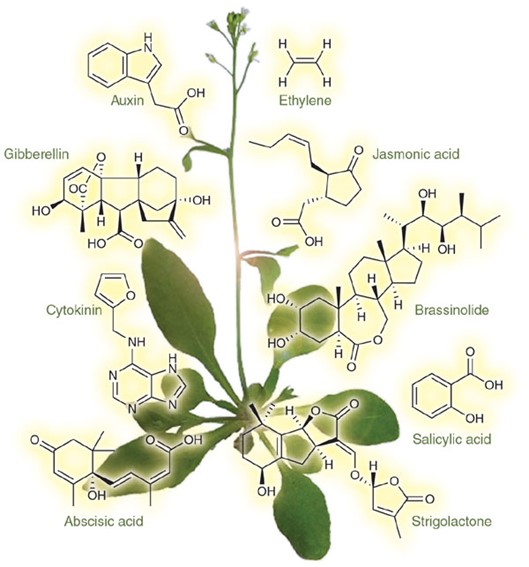
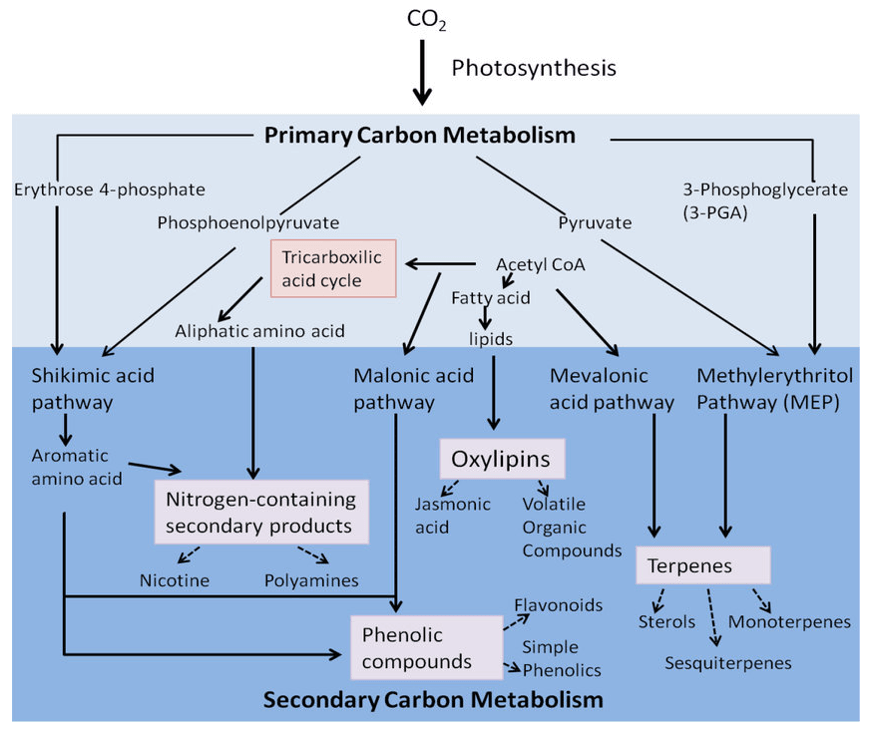
Plant Targeted Metabolomics: Precision for Plant Research
Plant targeted metabolomics is a specialized approach for accurately quantifying predefined plant metabolites using advanced LC-MS/MS platforms. Unlike untargeted metabolomics, which provides a broad but semi-quantitative overview, this method ensures absolute concentrations, high reproducibility, and pathway-specific coverage.
With this service, you can:
- Uncover stress-response mechanisms under drought, salinity, or pathogen attack.
- Support crop breeding programs by linking metabolites to yield and quality.
- Validate functional genomics findings through targeted metabolic profiles.
- Measure low-abundance compounds such as phytohormones with femtomolar sensitivity.
By focusing on the metabolites that matter most, our targeted metabolomics solution turns complex data into clear, actionable insights for research and development.
Why Use Plant Targeted Metabolomics?
Your plant-focused projects often demand more than a broad metabolic snapshot—they require pinpoint accuracy, clear biological context, and data you can rely on for confident decision-making. Our targeted metabolomics service meets those needs in five key ways:
- Pathway-Focused Confirmation
Measure only the metabolites that define your pathway of interest—phenylpropanoids, phytohormones, central-carbon intermediates, and more—so you can validate research hypotheses with precision.
- Ultra-Low Detection for Critical Signals
Triple-quadrupole LC-MS/MS in MRM mode reaches femtomolar sensitivity, enabling reliable quantification of ABA, SA, JA, and other signalling molecules that drive stress responses and developmental cues.
- Batch-to-Batch Reproducibility
Rigid QC checkpoints—pooled QC samples, isotope-labelled internal standards, and CV < 15 % across runs—ensure that the differences you observe between genotypes, treatments, or time points are truly biological.
- Actionable Markers for Breeding and Trait Selection
Customisable panels covering sugars, organic acids, amino acids, phenolics, and secondary metabolites deliver quantitative markers you can feed directly into QTL mapping, genomic-selection models, or quality-control pipelines.
- Transparent, Traceable Data Packages
Each report includes absolute concentrations, QC metrics, and instrument parameters, giving you complete traceability from sample to result—no hidden steps, no unanswered questions.
Targeted vs Untargeted Metabolomics: Quick Comparison
| Feature |
Plant Targeted Metabolomics |
Plant Untargeted Metabolomics |
| Purpose |
Quantification of selected metabolites |
Broad metabolite discovery |
| Output |
Absolute concentrations with QC validation |
Relative abundance values |
| Sensitivity |
High (femtomolar for phytohormones) |
Moderate |
| Best For |
Hypothesis-driven studies, regulated research |
Biomarker discovery, exploratory work |
Choose Targeted when accuracy, reproducibility, and specific pathway insights matter.
Why Choose Creative Proteomics for Plant Targeted Metabolomics?
- Comprehensive Metabolite Coverage
Analyze 300+ metabolites in one workflow—amino acids, organic acids, sugars, phenolics, alkaloids, and phytohormones.
- Ultra-Sensitive Detection
Triple-quadrupole LC-MS/MS in MRM mode achieves femtomolar limits of detection, ensuring reliable quantification of low-abundance signals.
- Uncompromising Quality Control:
- CV% ≤ 15% across replicates.
- Multi-point calibration with R² ≥ 0.99 for every analyte.
- Internal standards in every sample for matrix-effect correction.
- High-Throughput Capability
Advanced autosamplers handle up to 400 samples per batch, enabling large-scale or multi-location projects without data drift.
- Multi-Omics Integration
Combine metabolomics with transcriptomics or proteomics for system-level insights.
- Scalable for Big Projects
End-to-end SOPs ensure reproducibility across large breeding pipelines or multi-time-point studies.
Comprehensive Metabolite Coverage for Plant Research
Indoles and Indole-sulfur Compounds
Plant Targeted Metabolomics Technical Details & Coverage
At Creative Proteomics, precision begins with our instrumentation. We deploy a suite of triple-quadrupole LC-MS/MS systems—including SCIEX QTRAP® 6500+ and Waters Xevo TQ-S—optimized for Multiple Reaction Monitoring (MRM). This configuration guarantees selective detection and quantification of hundreds of plant metabolites in complex matrices.
Key technical highlights:
- Sensitivity: Detection down to femtomolar levels for low-abundance phytohormones and signaling molecules.
- Accuracy: Multi-point calibration curves with R² ≥ 0.99, ensuring reliable absolute quantification.
- Precision: Internal standards applied to every sample; CV% consistently ≤ 15% across technical replicates.
- Throughput: Automated UPLC systems and high-capacity autosamplers allow processing of up to 400 samples per batch, making it ideal for breeding pipelines and stress-response studies.
- Dynamic Range: Covers primary metabolites (amino acids, sugars) and secondary metabolites (flavonoids, alkaloids, phenolic acids) in a single analytical workflow.
Workflow Plant Targeted Metabolomics Service
Quality Assurance for Reliable Results
At Creative Proteomics, quality is embedded in every step of our plant targeted metabolomics workflow to ensure data that is not only accurate but reproducible and fully traceable.
Key QC Measures
- Internal Standard Normalization
Each sample includes isotope-labeled internal standards to correct for extraction variability and ion suppression.
- Calibration Accuracy
Multi-point calibration curves applied to every analytical batch with R² ≥ 0.99, guaranteeing precise absolute quantification.
- Instrument Stability Monitoring
QC samples injected after every 10 analyses to detect and correct for drift in real time.
- Stringent Data Acceptance Criteria
- CV% ≤ 15% across technical replicates.
- Back-calculated concentrations must fall within ±15% of expected values.
- Blank and recovery checks performed for every batch.
Result
Every dataset is validated through layered QC checkpoints, ensuring your conclusions are based on robust, reproducible, and decision-ready metabolite data.
Sample Requirements and Handling Guidelines
| Sample Type |
Minimum Amount |
Storage Condition |
Notes |
| Fresh Leaf Tissue |
≥ 500 mg |
Freeze in liquid N₂, store at -80°C |
Avoid degradation; no preservatives |
| Seeds / Grains |
≥ 200 mg |
Freeze in liquid N₂, store at -80°C |
Remove husks if applicable |
| Roots / Tubers |
≥ 500 mg |
Freeze in liquid N₂, store at -80°C |
Remove excess soil before freezing |
| Flowers / Fruits |
≥ 500 mg |
Freeze in liquid N₂, store at -80°C |
Separate tissue type if needed |
| Lyophilized Samples |
≥ 100 mg (dry weight) |
Store at -80°C |
Ensure samples are completely dried |
General Guidelines
- Avoid repeated freeze-thaw cycles.
- Submit samples in clearly labeled 2 mL cryotubes or sealed bags.
- Provide metadata (species, tissue type, treatment conditions) for accurate interpretation.
Applications: How Plant Targeted Metabolomics Accelerates Research
Deliverables: What You Get from Our Plant Targeted Metabolomics Service
Absolute Quantification Table
Accurate concentrations for all targeted metabolites, with QC metrics and calibration details.
Raw & Processed Files
Instrument files (.wiff, .mzML) and processed data tables (.xlsx) for easy downstream analysis.
Quality Control Documentation
Complete QC traceability, including internal standard correction, calibration curves (R² ≥ 0.99), and CV% summary.
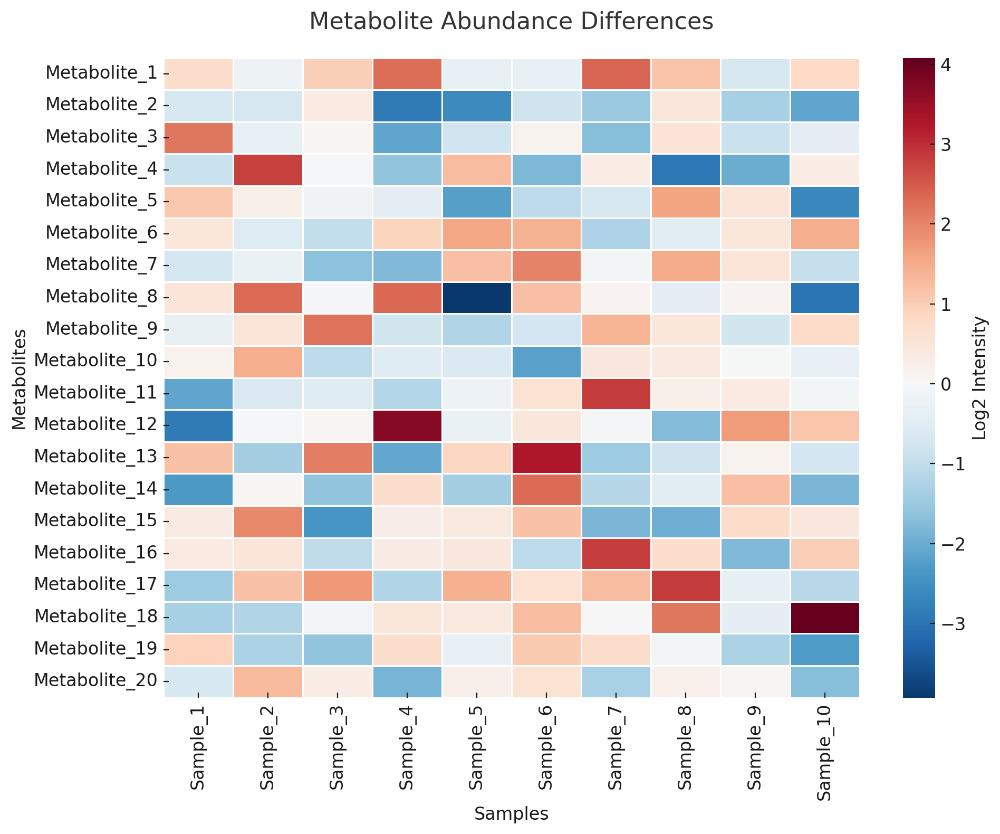
Visual Analytics Package (Optional)
- Heatmaps for comparative visualization
- PCA and OPLS-DA for clustering analysis
- KEGG pathway mapping for functional interpretation
Custom Reporting
Summary reports prepared for easy integration into your research workflow.
Integrated Analysis of Phytohormones Reveals Dynamic Responses to Salinity Stress in Arabidopsis thaliana Seedlings
Background
Understanding the molecular responses of plants to salinity stress is crucial for crop improvement. Phytohormones play a pivotal role in orchestrating these responses. This study aims to comprehensively analyze the dynamics of phytohormones in Arabidopsis thaliana seedlings under salinity stress.
Samples
Arabidopsis thaliana (Columbia-0 ecotype) seedlings were used for method validation and stress experiments. The study focused on shoots and roots harvested from plants subjected to either control conditions or 150 mM NaCl-induced salinity stress.
Technical Methods
Chemicals and Materials: Authentic standards and isotopically labeled counterparts were sourced from reputable suppliers. Chemicals, including formic acid, ACN, and MeOH, were procured from Merck.
Solubility Experiment: The solubility of selected BRs was investigated using different concentrations of aqueous ACN. UHPLC-ESI-MS/MS was employed for analysis, and relative yields were calculated.
Chlorophyll Extraction: Chlorophyll a and b were extracted using aqueous ACN at varying concentrations. Spectrophotometric measurements determined chlorophyll content.
Stability Experiment: Stability of analytes was examined under different conditions. Enzymatic activity in plant extracts was assessed using a proposed sample preparation protocol.
Sample Extraction and Purification: Plant material was extracted using ice-cold 50% aqueous ACN. Solid-phase extraction (SPE) with Oasis HLB RP cartridges facilitated purification.
UHPLC-ESI-MS/MS Conditions: Targeted compounds were analyzed using UHPLC coupled with a triple quadrupole mass spectrometer. Different elution conditions were applied for compounds detected in ESI(+) and ESI(-) modes.
Method Validation: Calibration curves, LOD, and LOQ were determined for UHPLC-ESI-MS/MS. Analyte losses during purification were evaluated, and method accuracy and precision were assessed.
Genevestigator Analysis: The Genevestigator tool was employed for meta-analytical assessment of gene expression under salt stress conditions, identifying salt stress as a significant modulator of hormone-related gene expression.
Results & Findings
Phytohormone Profiling: Comprehensive quantification of phytohormones provided insights into their dynamics under salt stress, highlighting key regulatory pathways.
Solubility Patterns: Solubility experiments revealed the behavior of selected BRs under different solvent conditions, aiding in the understanding of their chemical properties.
Chlorophyll Content: Chlorophyll extraction and spectrophotometric analysis provided information on the physiological impact of salt stress on Arabidopsis seedlings.
Stability of Analytes: Stability experiments elucidated the robustness of the proposed sample preparation protocol, ensuring reliable quantification of phytohormones.
Gene Expression Patterns: Genevestigator analysis corroborated the impact of salt stress on the expression of genes related to hormone biosynthesis and metabolism, aligning with the observed changes in phytohormone levels.
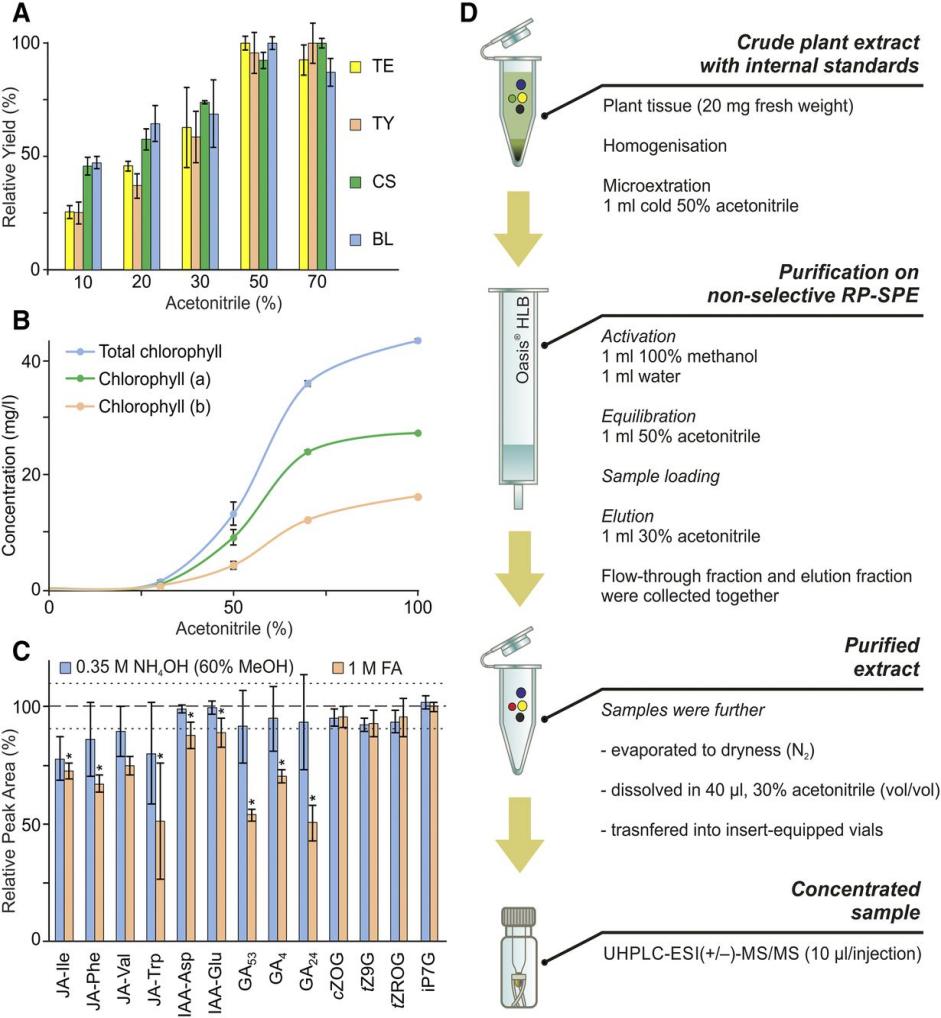 Optimization of sample preparation.
Optimization of sample preparation.
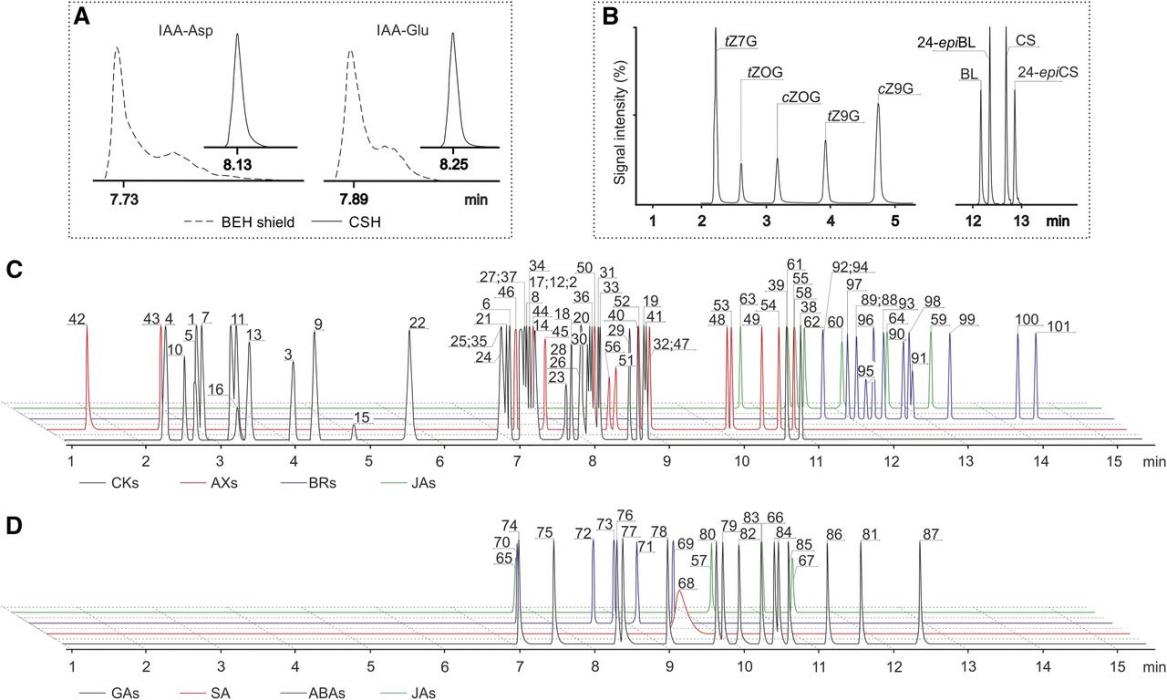 Optimization of baseline chromatographic separation.
Optimization of baseline chromatographic separation.
Reference
- Šimura, Jan, et al. "Plant hormonomics: multiple phytohormone profiling by targeted metabolomics." Plant physiology 177.2 (2018): 476-489.
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
Overexpression of maize ZmLOX6 in Arabidopsis thaliana enhances damage-induced pentyl leaf volatile emissions that affect plant growth and interaction with aphids
Tolley, J. P., Gorman, Z., Lei, J., Yeo, I. C., Nagashima, Y., Joshi, V., ... & Koiwa, H.
Journal: Journal of Experimental Botany
Year: 2023
DOI: https://doi.org/10.1093/jxb/erac522
Plant Growth Promotion, Phytohormone Production and Genomics of the Rhizosphere-Associated Microalga, Micractinium rhizosphaerae sp
Quintas-Nunes, F., Brandão, P. R., Barreto Crespo, M. T., Glick, B. R., & Nascimento, F. X.
Journal: Plants
Year: 2023
DOI: https://doi.org/10.3390/plants12030651
Summative and ultimate analysis of live leaves from southern US forest plants for use in fire modeling
Matt, F. J., Dietenberger, M. A., & Weise, D. R.
Journal: Energy & Fuels
Year: 2020
DOI: https://doi.org/10.1152/ajpgi.00184.2023
Disruption of CYCLOPHILIN 38 function reveals a photosynthesis-dependent systemic signal controlling lateral root emergence
Duan, L., Pérez-Ruiz, J. M., Cejudo, F. J., & Dinneny, J. R.
Journal: bioRxiv
Year: 2020
DOI: https://doi.org/10.1101/2020.03.11.985820
WI12 Rhg1 interacts with DELLAs and mediates soybean cyst nematode resistance through hormone pathways
Dong, J., & Hudson, M. E.
Journal: Plant Biotechnology Journal
Year: 2022
DOI: https://doi.org/10.1111/pbi.13709
Laboratory evaluation of larvicidal and oviposition deterrent properties of edible plant oils for potential management of Aedes aegypti (Diptera: Culicidae) in drinking water containers
Njoroge, T. M., & Berenbaum, M. R.
Journal: Journal of Medical Entomology
Year: 2019
DOI: https://doi.org/10.1093/jme/tjz021











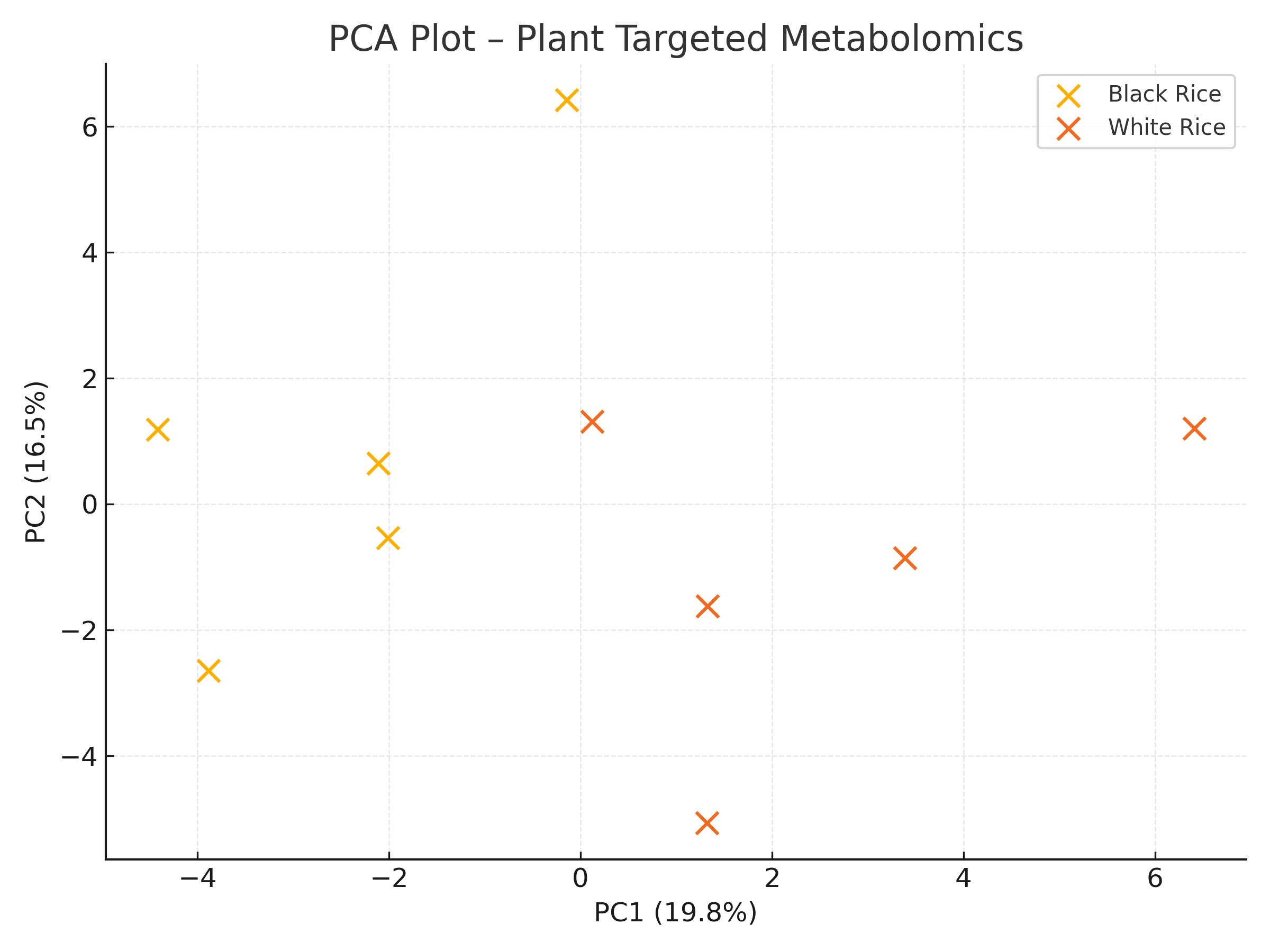
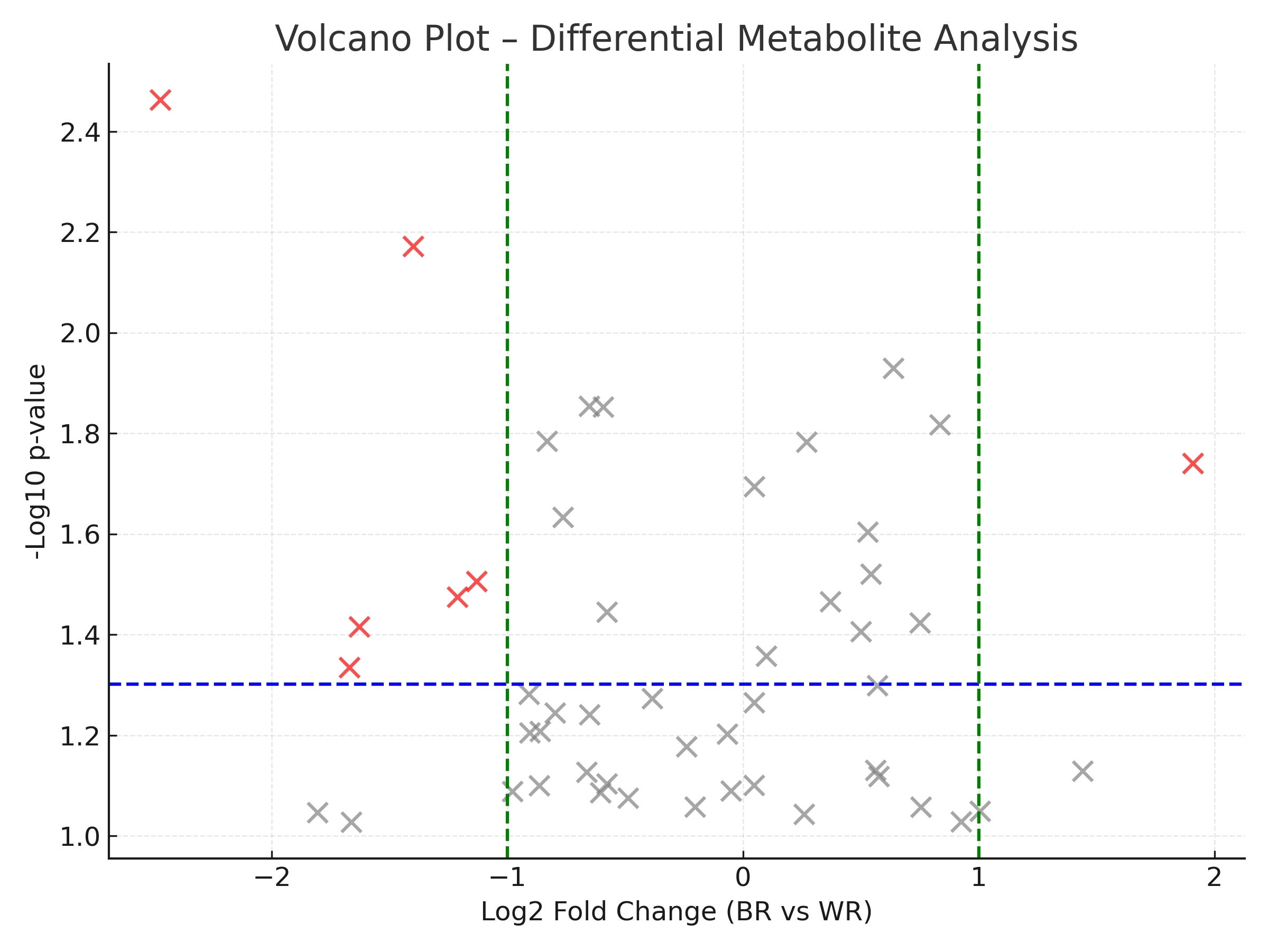
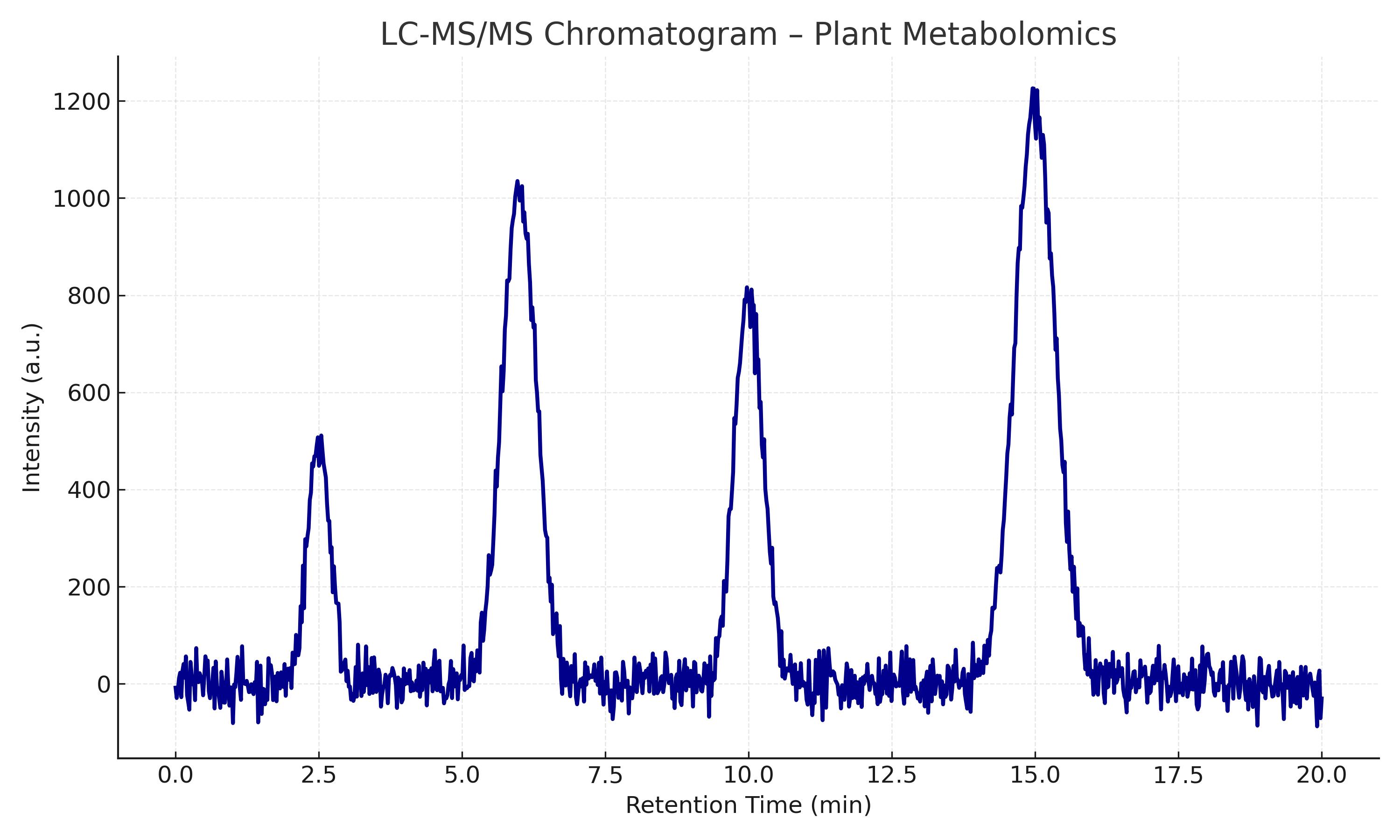
 Optimization of sample preparation.
Optimization of sample preparation. Optimization of baseline chromatographic separation.
Optimization of baseline chromatographic separation.