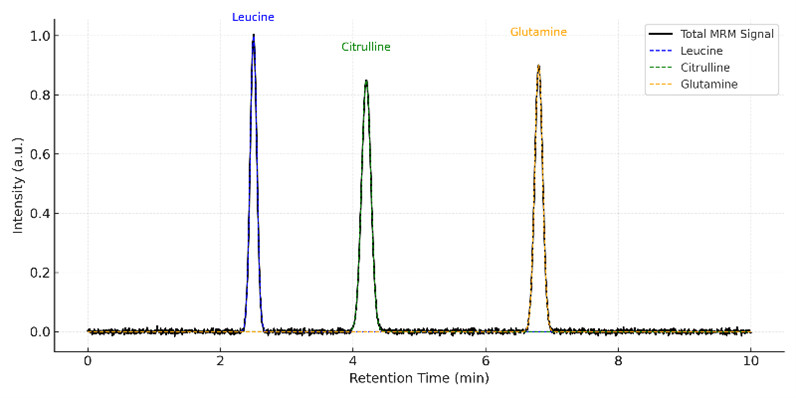
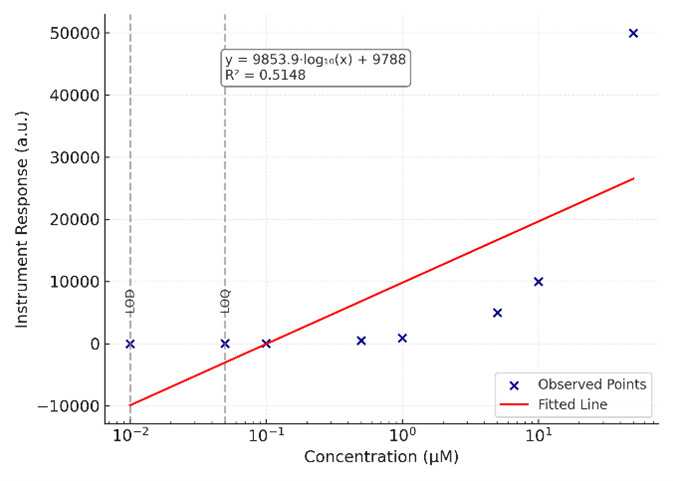
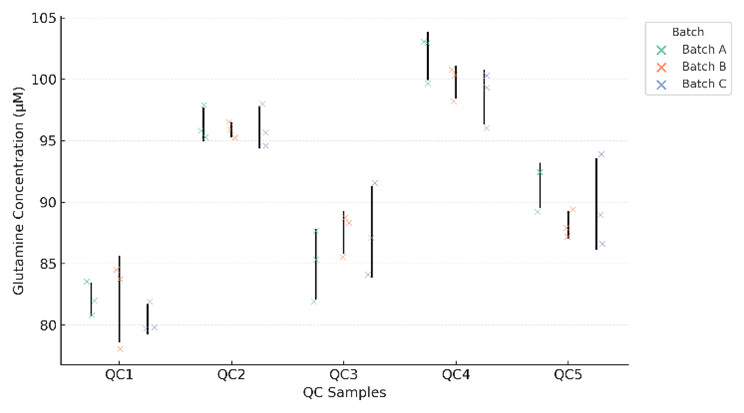
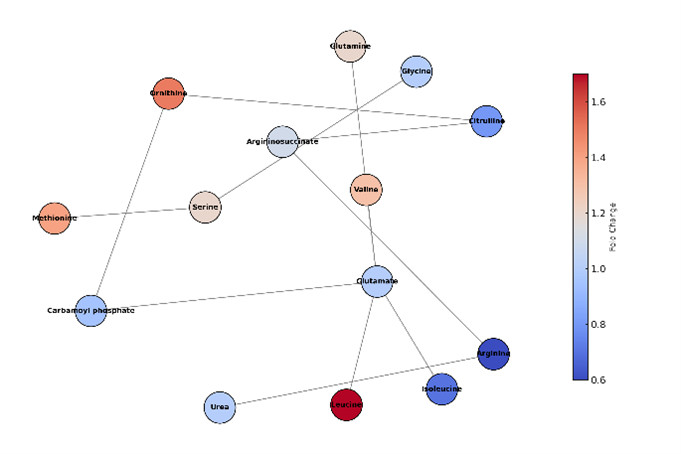
Amino Acid Metabolism — Why Targeted Quantification and Pathway Analysis Matter
Amino acid metabolism supports nitrogen balance, energy production, neurotransmitter synthesis, and protein turnover—core functions in both biological and engineered systems. These dynamic pools respond to nutrient inputs, process conditions, and metabolic interventions, making them critical readouts for pathway activity.
Targeted amino acid panel metabolomics via LC–MS/MS provides the most reliable approach for accurate amino acid quantification and pathway-level resolution of metabolic changes. It enables you to:
(i) measure absolute pool sizes and key metabolic ratios
(ii) monitor specific nodes such as the urea cycle , BCAA catabolism, one-carbon and sulfur amino acid pathways
(iii) compare conditions across biofluids, tissues, fermentation systems, food matrices, and plant materials.
What Problem Do We Solve?
Common obstacles include inconsistent quantification, matrix-driven ion suppression, and limited pathway coverage—all of which obscure signals and delay decisions. Creative Proteomics resolves these with a targeted service that is:
- Pathway-specific: Focused on quantifying amino acids that are central to metabolic networks such as the urea cycle, branched-chain amino acid metabolism, one-carbon metabolism, and neurotransmitter precursor pathways.
- Matrix-adapted: Compatible with diverse sample types—including biofluids, tissues, fermentation broths, food matrices, and plant extracts—with extraction and derivatization strategies tailored to each.
- Quantitatively reliable: Based on LC–MS/MS with isotope-dilution calibration, ensuring traceable, absolute concentration values, not just relative peak areas.
- QC-validated and reproducible: Every dataset includes bracketed calibrators, matrix-matched QCs, and system suitability metrics—so results are comparable across batches, projects, and labs.
Targeted Amino Acid Panels & Customization
Creative Proteomics offers a suite of LC–MS/MS-based targeted panels covering core amino acids, pathway-specific intermediates, and functional biomarkers across metabolic, nutritional, and neurobiological contexts. Panels are pre-configured or fully customizable with pathway relevance, matrix specificity, and quantification accuracy in mind.
Standard Quantification Panels
| Panel |
Included Analytes |
| Standard & Essential Amino Acids Panel (20 targets) |
Arginine, Histidine, Isoleucine, Leucine, Lysine, Methionine, Phenylalanine, Threonine, Tryptophan, Valine, Alanine, Asparagine, Aspartic acid, Cysteine*, Glutamine*, Glutamic acid, Glycine, Proline, Serine, Tyrosine |
| Comprehensive Amino Acid & Derivatives Panel (80+ targets) |
All core AAs plus:
L-4-Hydroxyproline, 5-Hydroxylysine, β-Alanine, 1-/3-Methyl-L-histidine, Citrulline, Ornithine, 4-Aminobutyric acid (GABA), Taurine, Monoamines, Catecholamines, Polyamines, Choline compounds, and other metabolic derivatives. |
* Cysteine, Glutamine, and Asparagine are stabilized through proprietary protocols to minimize degradation during sample handling and processing.
Pathway-Focused Panels
| Panel |
Key Targets / Pathways |
| Urea Cycle & Nitrogen Balance |
Arginine, Citrulline, Ornithine, Aspartate, Urea |
| Branched-Chain / Aromatic AAs (BCAA/AAA) |
Leucine, Isoleucine, Valine, Phenylalanine, Tyrosine, Tryptophan |
| One-Carbon & Sulfur Metabolism |
Methionine, Homocysteine, Serine, Glycine, Cysteine, Taurine |
| Neuroactive Precursors |
Glutamate, GABA, Aspartate, Tyrosine, Tryptophan |
| Amino Acid Transport / Biosynthesis |
Glutamine, Asparagine, Proline, Alanine, Threonine |
Functional Biomarker Panels
| Panel |
Representative Classes |
| Neurotransmitter-Related |
Monoamines, Catecholamines, GABA, Glutamate, Polyamines, Choline derivatives |
| Cytotoxicity Indicators |
Oxidized amino acid byproducts, 5-Hydroxylysine, β-Alanine, Methylhistidines |
| Immunological Biomarkers |
Arginine, Methionine, Kynurenine (customized on request) |
| Food Consumption Markers |
3-Methyl-L-histidine, β-Alanine, Taurine, Threonine, Glycine |
Customization & Add-ons
| Category |
Options |
| Target List |
Define your own list from amino acids, metabolic intermediates, amines, and custom biomarkers |
| Chiral Resolution |
Support for D/L isomer separation via derivatization or chiral LC |
| Internal Standards |
13C/15N-labeled internal standards for select analytes (quantitative or semi-quantitative) |
| Labile Compound Stabilization |
Special workflows for glutamine, cysteine, asparagine, and other unstable AAs |
Why Choose Our Amino Acid Metabolism Analysis Service?
- Curated Library of 150+ Metabolites
Our in-house MS/MS library covers over 150 amino acids and related compounds, enabling rapid panel setup, expansion, and confident annotation across core metabolic pathways.
- Up to 35 Isotope-Labeled Internal Standards
We apply pathway-balanced internal standards to control matrix effects and improve accuracy across structurally similar targets.
- Advanced Signal Processing
Data undergo isobar correction, ion ratio QC, and manual curation to ensure peak accuracy in complex matrices like fermentation broth or plant tissue.
- Batch-Reproducible & Method-Locked
Ideal for multi-batch studies, long-term process monitoring, and regulated environments—ensuring data comparability across time and scale.
Instrumentation & Method Parameters for Targeted Amino Acid Panel
Analytical Platform
LC–MS/MS (Primary Platform)
Mass Spectrometer: Triple Quadrupole MS (e.g., SCIEX QTRAP, Thermo TSQ, Agilent 6495 series)
Ionization Mode: Electrospray Ionization (ESI), Positive Mode
LC System: UHPLC or HPLC (depending on resolution and throughput needs)
Chromatography: HILIC or Reversed Phase, depending on analyte group and matrix
Derivatization Options: OPA, AccQ-Tag, FMOC-Cl, or derivatization-free (native) based on analyte class
Method Performance
| Parameter |
Typical Range |
| LOD / LOQ |
0.1–10 fmol (on-column), analyte-dependent |
| Dynamic Range |
4–5 orders of magnitude |
| Quantification Type |
Absolute (isotope dilution) and/or relative (normalized to internal standards) |
| Precision (CV%) |
≤10% intra-batch; ≤15% inter-batch |
| Accuracy (Spike Recovery) |
85–115% |
| Injection Volume |
Typically 2–10 μL |
GC–MS (Optional / Confirmatory Use)
GC–MS is available for specific cases requiring orthogonal confirmation, improved resolution, or library-based identification (e.g., β-alanine, GABA, short-chain amines).
- Derivatization: MSTFA (+1% TMCS), MTBSTFA, or ethyl chloroformate
- Column: DB-5ms or equivalent (30 m × 0.25 mm × 0.25 µm)
- Ionization: EI 70 eV, SIM or full-scan mode
- Typical Performance:
- LOD: 0.1–5 pmol
- Linearity: R² ≥ 0.995
- Precision: CV ≤ 10%
Internal Standards & Calibration
- Internal Standards: Up to 35 stable isotope-labeled AAs and analogs, selected by pathway grouping
- Calibration Strategy: 6–8 point standard curves, matrix-matched or surrogate matrix
- System Suitability: Ion ratio QC, retention time window, peak shape, carryover checks
Amino Acid Metabolism Analysis Workflow — A Step-by-Step Guide
How to Prepare and Submit Samples for Amino Acid Metabolomics
| Sample Type |
Amount Required |
Preparation Instructions |
Storage & Shipping |
| Plasma / Serum |
≥ 50 μL |
Collect in EDTA / heparin tube, centrifuge, aliquot supernatant, avoid hemolysis |
Store at −80°C; ship on dry ice |
| Urine / CSF |
≥ 50 μL |
Centrifuge to remove debris, aliquot |
Store at −80°C; ship on dry ice |
| Tissue (wet weight) |
5–20 mg |
Snap freeze in liquid nitrogen or dry ice immediately after collection |
Store at −80 °C; ship on dry ice |
| Cell Pellets |
≥ 0.5–5 × 106 cells |
Wash with cold PBS, centrifuge, remove supernatant, freeze pellet |
Store at −80°C; ship on dry ice |
| Fermentation Broth |
≥ 100 μL |
Centrifuge, collect supernatant or filter if needed |
Store at −80°C; ship on dry ice |
| Food / Ingredient |
50–100 mg or μL |
Homogenize or extract in cold solvent (e.g. methanol:water), aliquot |
Store at −80°C; ship on dry ice |
| Plant Tissue |
10–30mg |
Remove soil/debris, flash freeze immediately after collection |
Store at −80°C; ship on dry ice |
Notes
- Avoid repeated freeze-thaw cycles
- Clearly label each tube with sample ID, matrix type, date
- Submit sample list (Excel preferred) with matching metadata
- Contact us before sending rare, low-volume, or highly variable matrices
Deliverables: What You Receive from Amino Acid Metabolomics Analysis
- Quantification tables:absolute and/or relative amino acid concentrations (.csv / Excel)
- Calibration & QC report:calibration curves, accuracy, precision, recovery, carryover checks
- Raw & processed data:instrument raw files, processed peak lists, chromatograms
- Method documentation:LC–MS/MS parameters, derivatization protocol, internal standard setup
- Pathway annotations:KEGG / HMDB / CAS IDs linked to quantified metabolites
- Final summary report:sample notes, QC results, interpretation highlights
Applications of Amino Acid Profiling
Our amino acid profiling service supports researchers and developers across industries:
Drought-Stressed Switchgrass: Water-Soluble Saponins Inhibit Yeast Growth
Background
Researchers working with switchgrass hydrolysates observed complete inhibition of yeast growth in material harvested under drought conditions. To diagnose the cause, biomass was pretreated via Ammonia Fiber Expansion (AFEX), subjected to solvent extraction (water, ethanol, ethyl acetate) either before or after AFEX, and then analyzed by liquid chromatography–mass spectrometry (LC–MS) while monitoring Saccharomyces cerevisiae fermentation performance.
Challenge:
Identify the plant-generated vs. pretreatment-derived inhibitors in a complex plant matrix, and determine a practical mitigation that restores fermentation.
Findings (from the published study)
- Water extraction before AFEX alleviated the inhibition of yeast fermentation observed for drought-year switchgrass.
- Saponins were significantly more abundant in water extracts from the drought-year biomass; the team highlighted features at m/z corresponding to 1176 and 1212 Da, both sharing the diosgenin aglycone.
- Add-back tests using protodioscin (a commercial saponin) reproduced yeast growth inhibition in otherwise non-inhibitory hydrolysate.
- The authors report up to ~16-fold higher saponins under drought stress and conclude that water-soluble saponins likely contribute to the inhibitory phenotype.
Table 4A. Amino acid composition in hydrolysates of ethanol extracted switchgrass
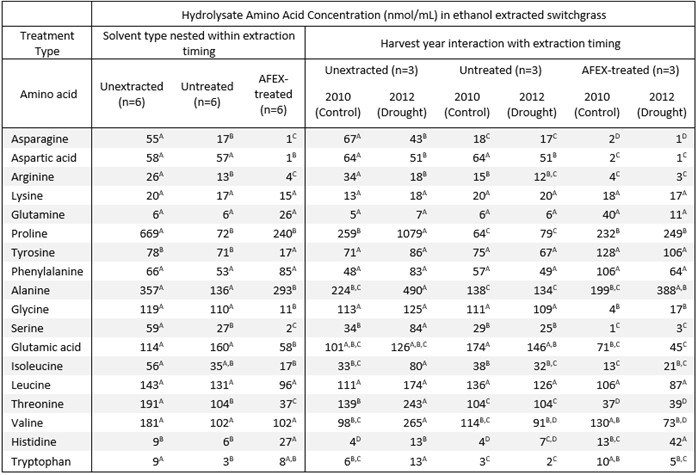
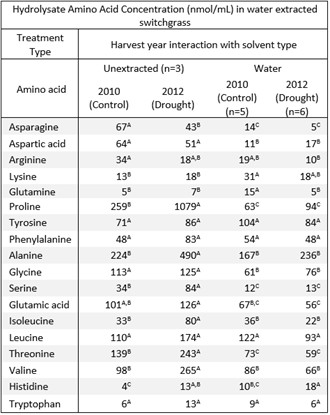
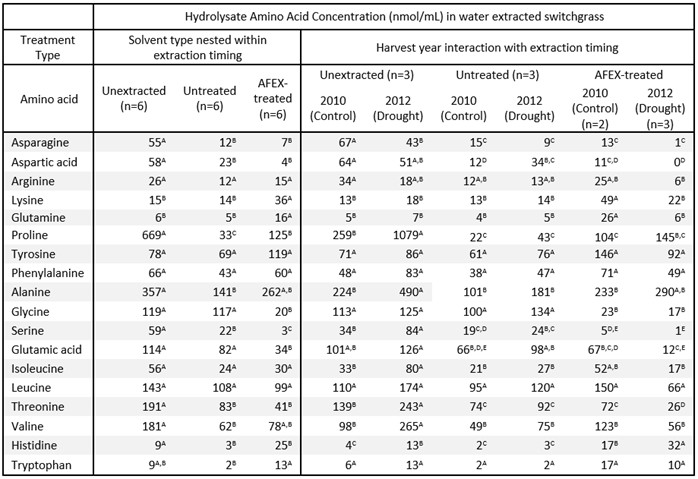
Table 4B. Amino acid composition in hydrolysates of water extracted switchgrass
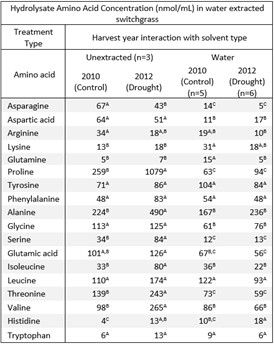
Process Insight
The study demonstrates a feedstock-QA workflow that combines water extraction, AFEX, high-solids enzymatic hydrolysis, LC–MS characterization of extracts, and S. cerevisiae performance readouts to pinpoint inhibitory chemistry under drought stress.
Where Our Targeted Amino Acid Panel Fits
While this publication pinpoints saponins as key inhibitors, Creative Proteomics supports similar bioenergy projects by integrating amino acid analysis alongside saponin surveillance to deliver nitrogen-pathway context that impacts fermentation:
- Hydrolysate & broth amino acid profiling to track free amino acids, BCAA levels, and urea-cycle intermediates that influence yeast nitrogen assimilation and growth kinetics.
- LC–MS/MS (MRM) with isotope-dilution for absolute quantification and cross-batch comparability—designed to slot into the same AFEX–hydrolysis–fermentation pipeline reported in the study.
- Pathway-linked outputs (KEGG/HMDB IDs) to interpret amino-nitrogen status in tandem with water-soluble inhibitor trends
Reference
- Chipkar, S., Smith, K., Whelan, E.M. et al. Water-soluble saponins accumulate in drought-stressed switchgrass and may inhibit yeast growth during bioethanol production. Biotechnol Biofuels 15, 116 (2022).
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566