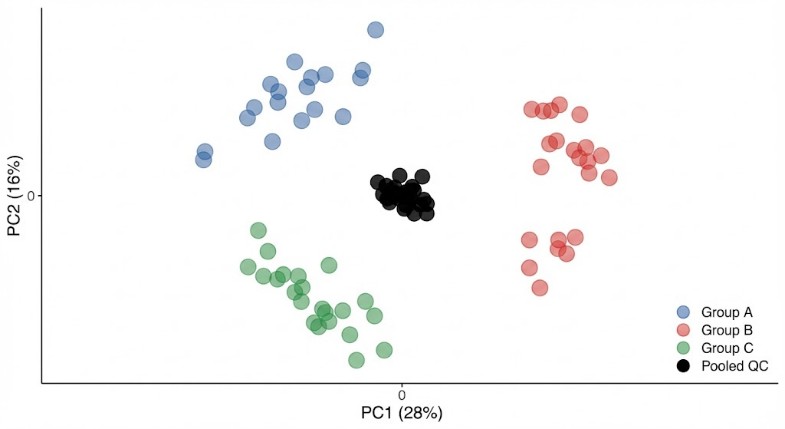
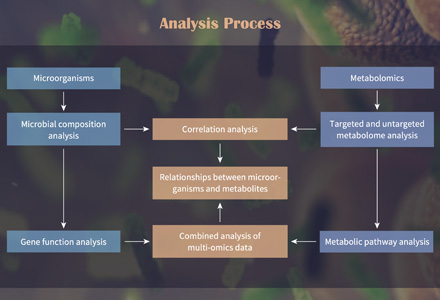
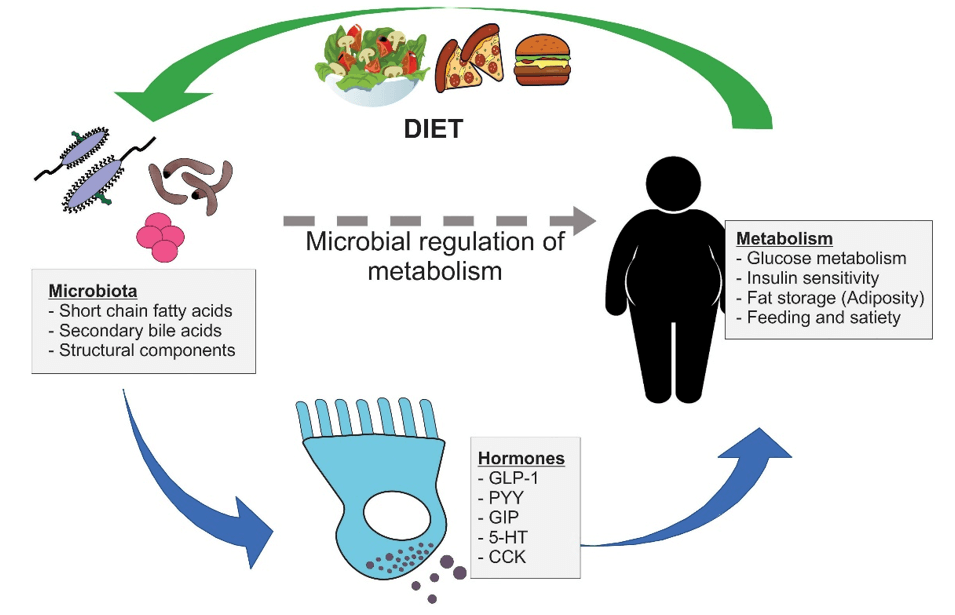
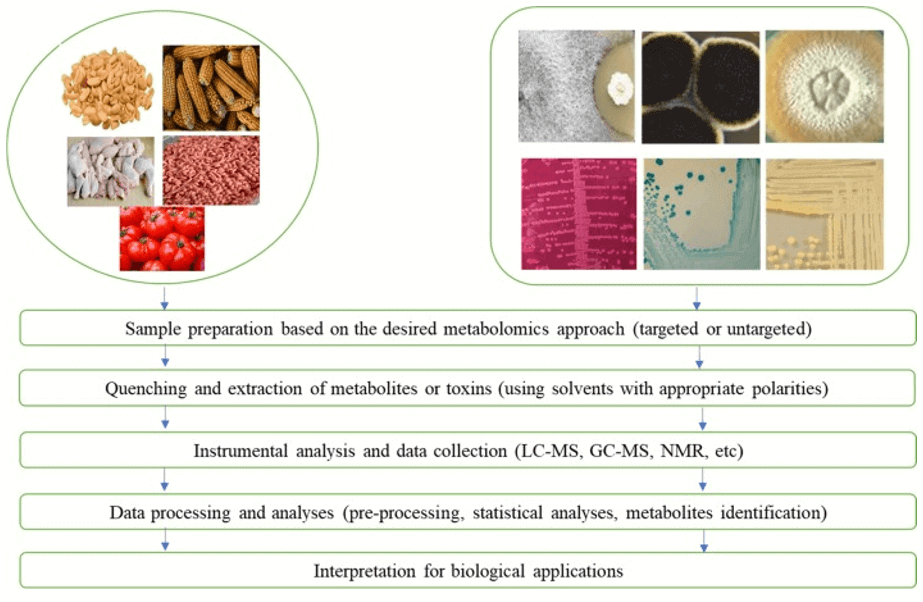
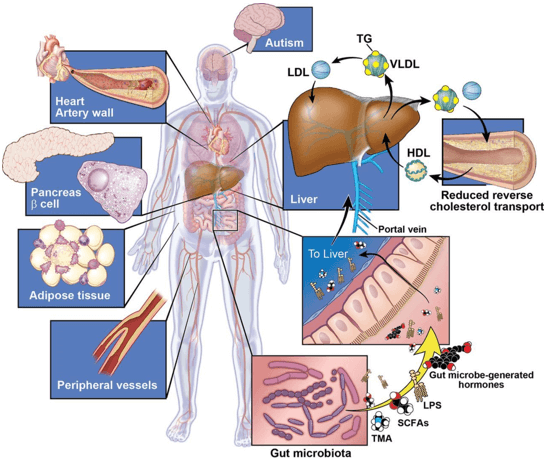
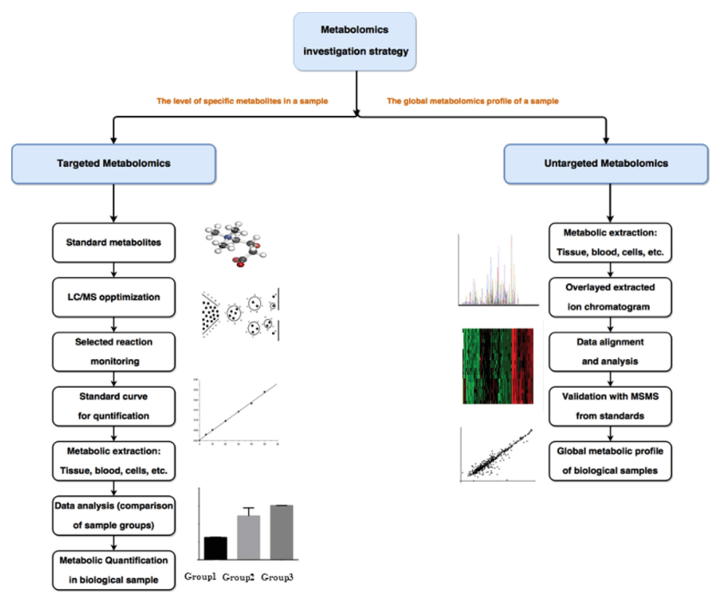
| SCFAs |
Acetic acid (acetate), Propionic acid (propionate), Butyric acid (butyrate), Isobutyric acid, Valeric acid, Isovaleric acid, Caproic acid (hexanoate) |
Targeted (Absolute) |
GC–MS |
| Organic Acids & Central Carbon Intermediates |
Lactate, Pyruvate, Succinate, Malate, Fumarate, Citrate, α-Ketoglutarate, Oxaloacetate, Formate |
Targeted / Untargeted |
LC–MS |
| Bile Acids (Primary/Secondary) |
Cholic acid (CA), Chenodeoxycholic acid (CDCA), Deoxycholic acid (DCA), Lithocholic acid (LCA), Ursodeoxycholic acid (UDCA), Hyodeoxycholic acid (HDCA) |
Targeted (Absolute) / Profiling |
LC–MS/MS (MRM) |
| Bile Acids (Conjugated) |
Taurocholic acid (TCA), Glycocholic acid (GCA), Taurochenodeoxycholic acid (TCDCA), Glycochenodeoxycholic acid (GCDCA), Taurodeoxycholic acid (TDCA), Glycodeoxycholic acid (GDCA), Taurolithocholic acid (TLCA), Glycolithocholic acid (GLCA), Tauro-UDCA (TUDCA), Glyco-UDCA (GUDCA) |
Targeted (Absolute) / Profiling |
LC–MS/MS (MRM) |
| Tryptophan Pathway (Indoles & Related) |
Tryptophan, Indole, Indole-3-acetic acid (IAA), Indole-3-propionic acid (IPA), Indole-3-lactic acid (ILA), Indole-3-aldehyde (IAld), Skatole (3-methylindole), Tryptamine |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Kynurenine Pathway |
Kynurenine, Kynurenic acid, 3-Hydroxykynurenine, Quinolinic acid |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Serotonin-Related (Project-dependent) |
Serotonin (5-HT), Melatonin |
Targeted (Absolute) / Relative |
LC–MS/MS |
| Phenylalanine/Tyrosine-Derived Microbial Metabolites |
Phenylalanine, Tyrosine, Phenylacetic acid (PAA), Phenylpropionic acid (PPA), Phenyllactic acid (PLA), p-Cresol, p-Cresyl sulfate (project-dependent), 4-Hydroxyphenylacetic acid, 4-Hydroxyphenyllactic acid, Homovanillic acid (project-dependent) |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Choline–Carnitine–TMA Axis |
Choline, Betaine, Carnitine, Trimethylamine (TMA, project-dependent), Trimethylamine N-oxide (TMAO), γ-Butyrobetaine |
Targeted (Absolute) |
LC–MS/MS (MRM) |
| Amino Acids (BCAA/AAA & Others) |
Leucine, Isoleucine, Valine; Phenylalanine, Tyrosine, Tryptophan; Alanine, Glycine, Serine, Threonine, Methionine, Lysine, Arginine, Histidine, Proline, Glutamate, Glutamine, Aspartate, Asparagine, Cysteine |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Biogenic Amines (Project-dependent) |
GABA, Putrescine, Cadaverine, Spermidine, Spermine, Histamine |
Targeted (Absolute) / Relative |
LC–MS/MS |
| Neuroactive/Bioactive Metabolites (Project-dependent) |
GABA, Dopamine (project-dependent), Norepinephrine (project-dependent), Acetylcholine (project-dependent), Nicotinic acid (Niacin), Nicotinamide |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Vitamins/Cofactors v(Project-dependent) |
Riboflavin (B2), Pyridoxal phosphate (B6), Pantothenate (B5), Biotin (B7) |
Targeted / Untargeted |
LC–MS/MS / HRMS |
| Lipidomics (Optional module; class-level coverage) |
Phosphatidylcholines (PC), Phosphatidylethanolamines (PE), Phosphatidylserines (PS), Phosphatidylinositols (PI), Sphingomyelins (SM), Ceramides (Cer), Triacylglycerols (TAG), Diacylglycerols (DAG), Free fatty acids (FFA), Acylcarnitines, Lysophospholipids (LPC/LPE) |
Lipidomics (Profiling) |
LC–MS (HRMS/MSMS) |