What Are Carotenoids and Why Analyze Them?
Carotenoids are key pigments that drive plant coloration, photoprotection, and nutritional quality. Creative Proteomics provides a dedicated carotenoids analysis service based on a 66-compound targeted metabolomics panel covering carotenes, xanthophylls, and carotenoid esters. The workflow is optimized for plant tissues, foods, feeds, and algal biomass, delivering robust quantitative profiles that can be directly used in breeding, nutrition, and functional food research.
Why Quantify Carotenoids in Plant and Food Research?
Accurate carotenoid measurements turn visible traits and quality attributes into quantitative endpoints. In many projects, carotenoid data are used to:
- Relate pigment composition to fruit, grain, and leaf coloration.
- Evaluate provitamin A and xanthophyll levels as part of nutritional quality assessment.
- Track carotenoid changes under stress, storage, and processing conditions.
- Provide biochemical readouts that complement genomic, transcriptomic, and phenotype data.
By combining targeted carotenoid profiling with your existing datasets, you can move from descriptive color observations to mechanistic and data-driven decisions.
Carotenoids Analysis Service: Project Types and Use Cases
Project Types We Commonly Support
Carotenoid profiling needs can look very different from one project to another. In practice, most studies that use our carotenoids panel fall into a few broad types:
You do not need to fit perfectly into one category. We use these project types as a starting point to adapt the carotenoid panel and sampling strategy to your specific questions.
Questions This Service Is Designed to Answer
Rather than offering "just another assay," this service is structured to answer research and development questions such as:
- Which lines or treatments show the highest levels of key carotenes or xanthophylls?
- How do pigment profiles shift across developmental stages, stress conditions, or storage time points?
- Are carotenoid changes aligned with transcript-level regulation in biosynthetic or degradation pathways?
- Does a new processing or formulation strategy preserve, enhance, or degrade carotenoids compared with a control?
- Can we distinguish between free and esterified carotenoids where that distinction is biologically or nutritionally relevant?
By framing the study around these kinds of questions, we can help you decide which analytes, time points, and sample types need to be prioritized.
How Our Panel Fits Into Your Study Design
The carotenoids panel is most effective when it is integrated into a broader experimental plan, not run in isolation. Typical ways our clients use the panel include:
- As a primary endpoint
Using quantitative carotenoid profiles as the main readout to rank lines, treatments, or storage conditions.
- As a mechanistic layer
Overlaying carotenoid data onto transcriptomics, genomics, or physiological measurements to clarify how pigment pathways contribute to observed traits.
- As a quality and consistency check
Incorporating carotenoid measurements into routine QC panels for raw materials, intermediates, or pilot-scale batches in research settings.
During the initial discussion, we map your study design onto these roles and confirm how carotenoid data will be interpreted alongside your other readouts. This ensures that the panel is configured to support decisions, not just to generate additional numbers.
Carotenoid Panel Coverage: 66 Compounds Across Key Classes
| Class |
Compounds |
| Carotenes |
Alpha-Carotene; Beta-Carotene; Gamma-Carotene; Lycopene; Phytofluene; (E/Z)-Phytoene; Epsilon-Carotene |
| Xanthophylls |
Antheraxanthin; Zeaxanthin; Violaxanthin; Neoxanthin; Lutein; Beta-Cryptoxanthin; Astaxanthin; Apocarotenal; Capsanthin; Alpha-Cryptoxanthin; Capsorubin; Canthaxanthin; Echinenone; Beta-Citraurin |
| Carotenoid esters |
Lutein Caprate; Lutein Laurate; Lutein Myristate; Lutein Palmitate; Lutein Stearate; Lutein Dilaurate; Lutein Dipalmitate; Lutein Distearate; Zeaxanthin Dipalmitate; Zeaxanthin Dilaurate; Zeaxanthin-Laurate-Myristate; Zeaxanthin Dimyristate; Zeaxanthin-Laurate-Palmitate; Zeaxanthin-Myristate-Palmitate; Violaxanthin Laurate; Violaxanthin Myristate; Violaxanthin Palmitate; Violaxanthin Dilaurate; Violaxanthin Dimyristate; Beta-Cryptoxanthin Laurate; Beta-Cryptoxanthin Myristate; Beta-Cryptoxanthin Palmitate; Beta-Cryptoxanthin Oleate; Rubixanthin Laurate; Rubixanthin Myristate; Rubixanthin Palmitate |
For carotenoid esters, only key representatives are shown here. If you need the complete panel list or want to confirm coverage for specific targets, please contact our team, and we will provide detailed information tailored to your project.
Why Choose Creative Proteomics for Carotenoid Analysis?
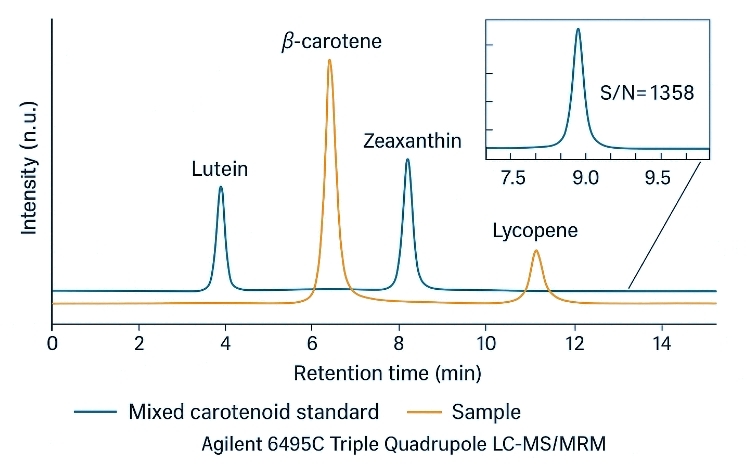
- Sensitive quantification: APCI-MRM detects many carotenoids at low-ng/mL levels.
- High chromatographic resolution: UHPLC gradients resolve >60 carotenoids, including critical isomer pairs.
- Comprehensive 66-compound panel: Covers carotenes, xanthophylls, and esters in one run.
- Strong reproducibility: Typical intra-batch RSD <10% for key analytes.
- Accurate calibration: Matrix-aware curves with R² ≥ 0.99 across working ranges.
- Degradation-protected extraction: Low-light, low-temperature workflow preserves true carotenoid ratios.
Analytical Platforms for Carotenoid Quantification
For carotenoid analysis, we use triple-quadrupole LC–MS/MS platforms (e.g., AB Sciex QTRAP 6500 or equivalent) with APCI and targeted MRM acquisition, coupled to UHPLC separation optimized for plant and food matrices.
When UV–Vis information or alternative detection is helpful, LC–MS/MS can be complemented by HPLC-DAD or HPLC-MS methods for additional confirmation and method flexibility.
Method Performance Parameters
| Parameter |
Typical configuration / performance (carotenoid method) |
| Instrument platform |
Triple-quadrupole LC–MS/MS (e.g., AB Sciex QTRAP 6500 or equivalent) |
| Chromatographic resolution |
UHPLC reversed-phase gradient optimized to separate major carotenes and xanthophylls; critical pairs verified in method validation |
| Transitions per compound |
1–2 optimized MRM transitions per carotenoid, selected for specificity and robustness |
| Limit of Detection (LOD) |
Low ng/mL to low pmol range for many carotenoids in typical plant/food matrices (compound- and matrix-dependent) |
| Limit of Quantification (LOQ) |
Defined per compound based on signal-to-noise and precision criteria within the validated range |
| Linearity range |
External calibration with internal standards; linear fit with typical R² ≥ 0.99 over the project-specific range |
| Method precision (RSD) |
Intra-batch RSD (peak area or concentration) controlled within commonly accepted targeted metabolomics QC criteria |
| Recovery rate |
Evaluated using spiked or matrix-matched samples; acceptance ranges set according to project requirements and compound class |
| Calibration approach |
Matrix-aware external calibration supported by isotope-labeled or structurally related internal standards |
| QC strategy |
Pooled QC samples, system suitability checks, and repeat injections to monitor RT stability, response drift, and batch-to-batch consistency |
If you need to align with a specific internal SOP, guideline, or journal requirement, we can provide project-level method details—such as target LOD/LOQ, linearity range, recovery acceptance windows, and QC criteria—during study design and quotation.
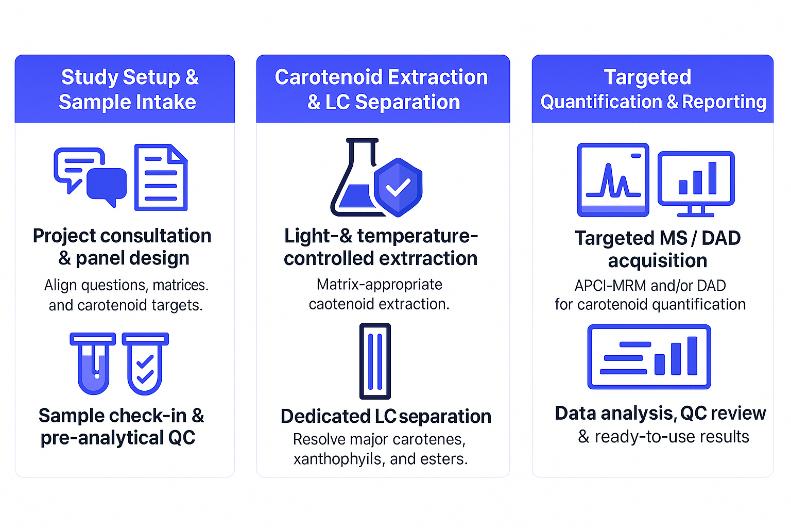
From Sample to Report: Workflow for Carotenoid Profiling Projects
Sample Requirements and Submission Guidelines for Carotenoid Analysis
| Sample type |
Recommended amount* |
Container |
| Fresh plant leaves / needles |
≥ 0.3 g fresh weight per sample |
2 mL or 5 mL screw-cap tube |
| Fresh fruits / fleshy tissues (pericarp, pulp) |
≥ 0.5 g fresh weight per sample |
5 mL or 15 mL screw-cap tube |
| Roots / tubers |
≥ 0.5 g fresh weight per sample |
5 mL or 15 mL screw-cap tube |
| Seeds / grains (whole seeds) |
≥ 0.5 g |
2 mL or 5 mL screw-cap tube |
| Ground grain flour / meal |
≥ 0.3 g dry powder |
2 mL or 5 mL screw-cap tube |
| Lyophilized plant tissues / powders |
≥ 0.2 g dry weight |
2 mL or 5 mL screw-cap tube |
| Processed foods (puree, juice, paste) |
≥ 1 mL or 1 g |
5 mL or 15 mL screw-cap tube |
| Processed foods (powders, snacks, cereals) |
≥ 0.5 g dry weight |
2 mL or 5 mL screw-cap tube |
| Oils / lipid extracts |
≥ 0.5 mL |
Amber glass vial with tight cap |
| Algal biomass / wet pellets |
≥ 0.3 g wet weight |
2 mL or 5 mL screw-cap tube |
*If your available sample amount is lower than recommended, please contact us in advance so we can evaluate feasibility and adjust the workflow where possible.
General notes for sample collection and submission
- Avoid light and heat
Carotenoids are light- and temperature-sensitive. Collect and handle samples as quickly as possible, keep them cold and shielded from light (e.g., using aluminum foil or amber containers).
- Do not add preservatives or extraction solvents
Unless agreed in advance, please send untreated biological samples (no buffers, detergents, antioxidants or organic solvents added). If you have already extracted pigments, provide details of the extraction protocol.
- Unsure about your sample type?
If your matrix is not listed in the table (e.g., specific processed products, mixed samples, unusual tissues), you can contact us with a short description. We will confirm sample amount and handling instructions before shipment.
Final Deliverables: Carotenoid Quantification Report
Quantitative data table: Excel/CSV file with carotenoid concentrations for all samples, including sample IDs and basic group/replicate information.
Method summary: Brief PDF/text description of sample preparation, HPLC conditions, LC–MS/MS settings and reporting units.
QC summary: Key quality control metrics (e.g., calibration range and linearity, QC sample precision) for the batch.
Applications of Carotenoid Metabolomics in Plant & Food Research
How to Design a Successful Carotenoids Metabolomics Study
Choosing Sample Types, Time Points, and Controls
Good carotenoid study design starts with sampling decisions:
- Align time points with biological events such as ripening, stress exposure, or processing steps.
- Include the most relevant tissues (e.g., fruit flesh, peel, grain endosperm) and, where useful, reference tissues.
- Define clear controls and baselines so that carotenoid shifts can be interpreted in context.
Share your planned sampling scheme with us and we can highlight gaps or refinements before you commit material.
Combining Carotenoids with Other Metabolite Panels
Carotenoids rarely act in isolation. For many projects it is useful to:
- Pair carotenoid profiling with broader lipidomics, pigmentomics, or primary metabolite panels.
- Harmonize sample preparation across panels to reduce variability and rework.
- Plan in advance how carotenoid, metabolite, transcript, and phenotype data will be integrated.
Our team can suggest compatible panel combinations and help you prioritize analytes around your main research questions.
Common Pitfalls and How We Help You Avoid Them
Typical issues that undermine carotenoid data quality include:
- Light and temperature exposure – carotenoids degrade when sampling, transport, or extraction are not protected.
- Insufficient material or replication – marginal amounts and too few biological replicates weaken conclusions.
- Uncontrolled batch effects – mixing groups across analytical batches without adequate QC introduces bias.
By engaging early, we can help you structure sampling, storage, batching, and QC so that carotenoid datasets remain robust and aligned with your statistical analysis plan.
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024