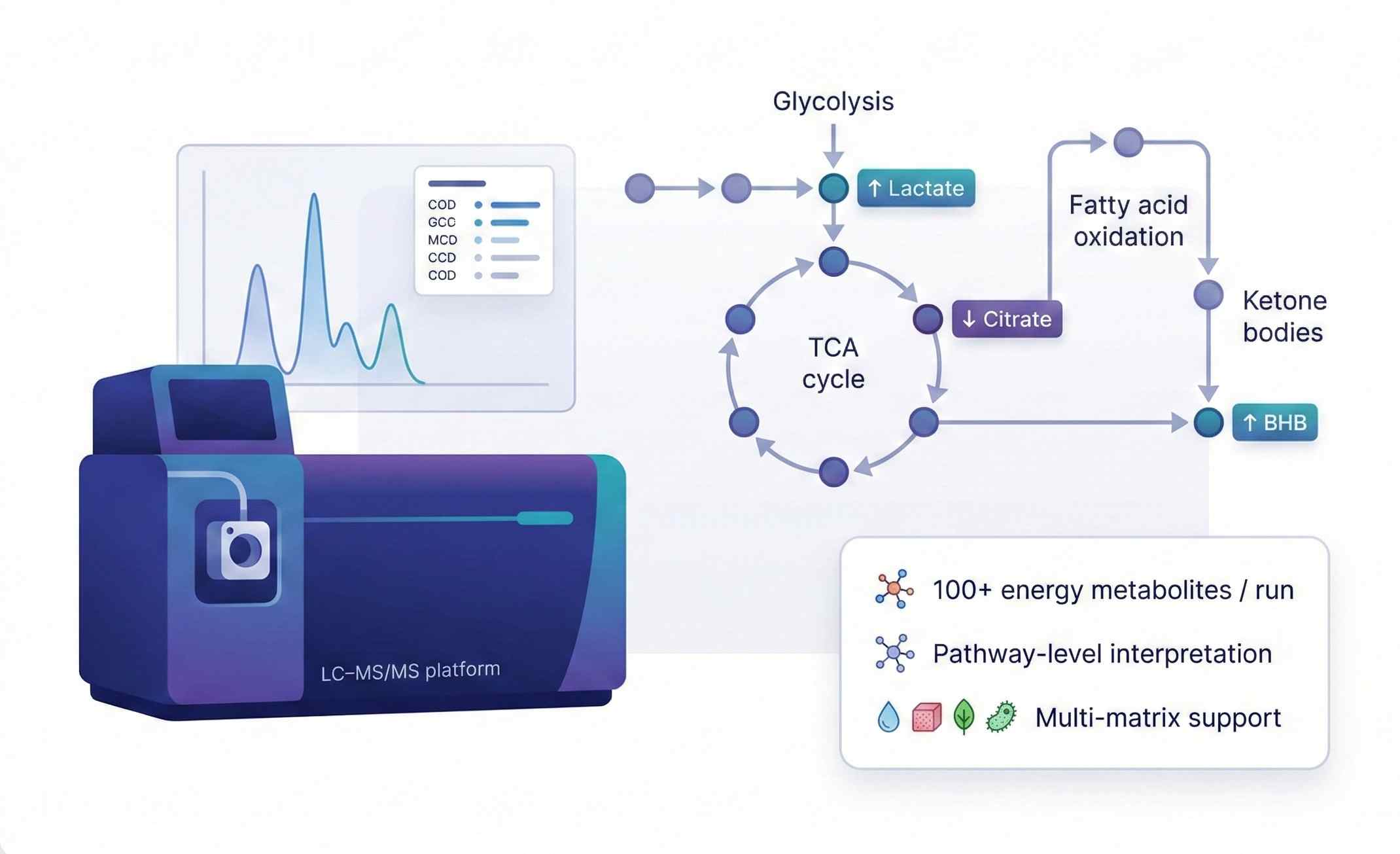
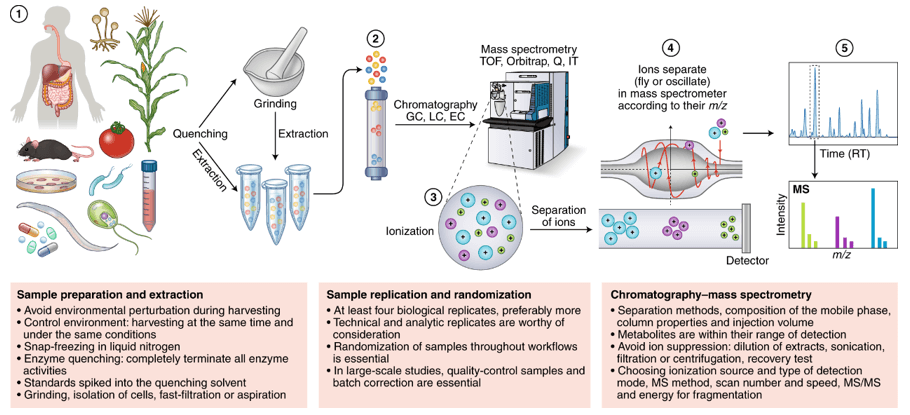
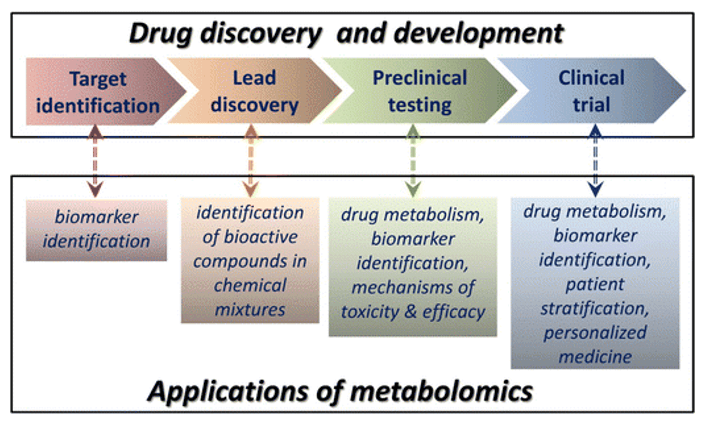
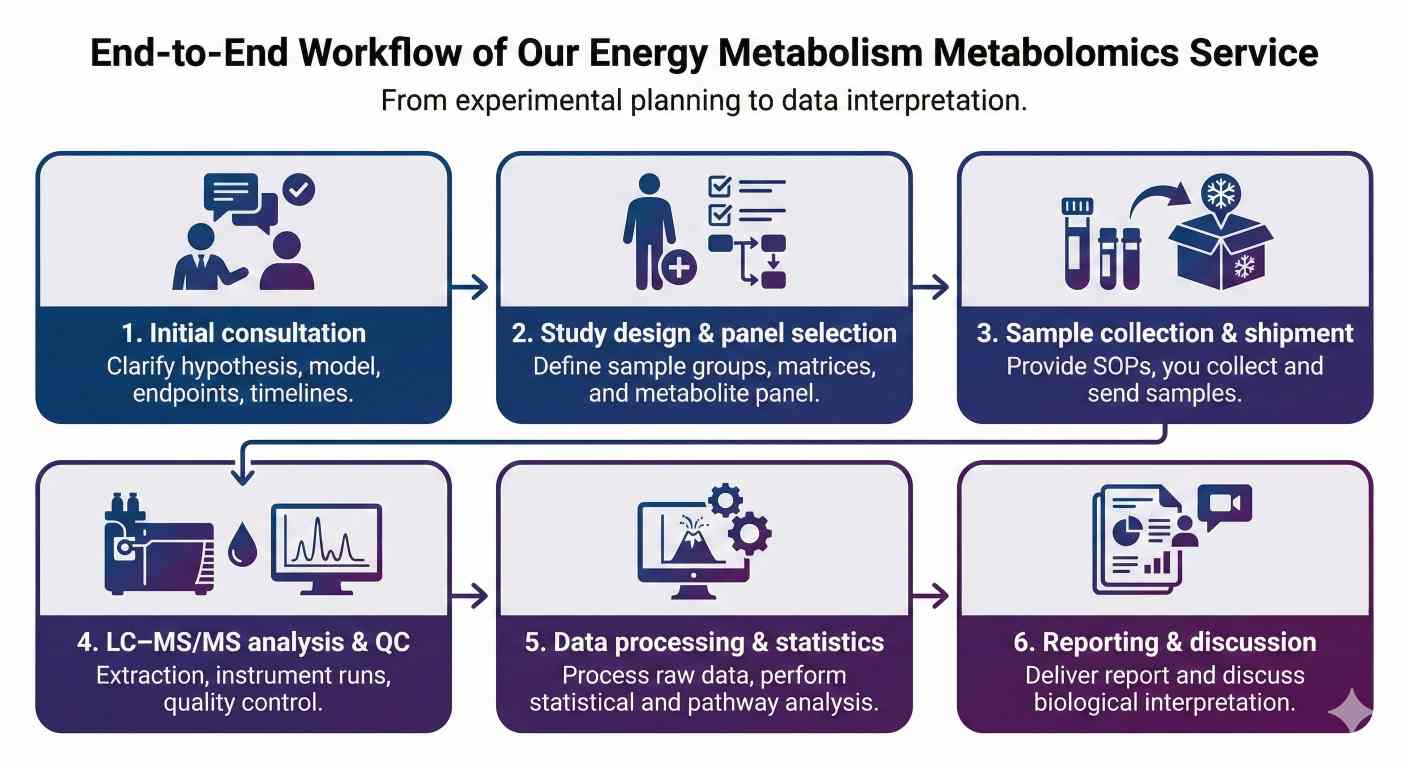
Overview of Our Energy Metabolism Metabolomics Service
Energy metabolism is central to how cells grow, adapt, and respond to stress. It links:
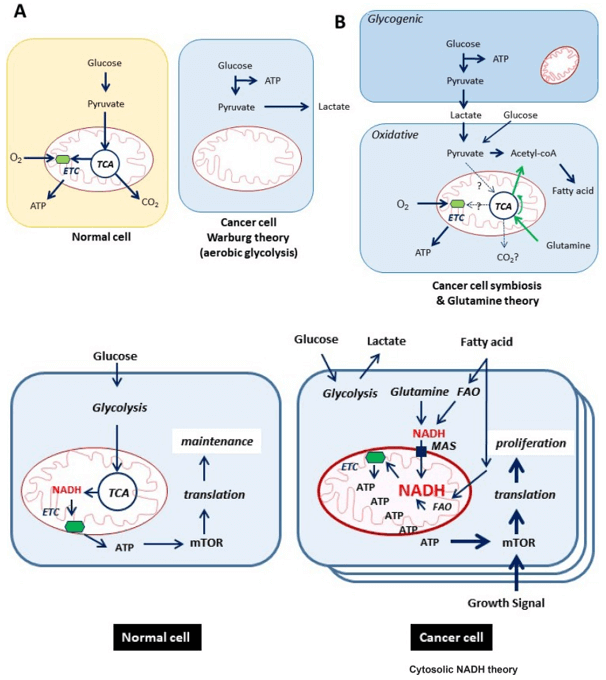
- Carbon flux through glycolysis and the TCA cycle
- Mitochondrial function and oxidative phosphorylation
- Amino acid utilization, lipid oxidation, and ketone body production
Our energy metabolism metabolomics service provides targeted LC–MS/MS panels together with broader LC–MS-based metabolomics profiling focused on these pathways. It is designed to help you:
- Demonstrate how a gene, drug, diet, training program, or stress condition reshapes energy metabolism
- Connect phenotypes, transcriptomics, and proteomics with metabolite-level changes
- Build a clear mechanistic understanding of how energy pathways are regulated in your model
Metabolites and Pathways Covered
We focus on key nodes that define cellular and whole-body energy status. Panels can be customized to your model and research question.
Key Pathways and Example Metabolites
| Pathway / Module |
Example Metabolites* |
Typical Questions Addressed |
| Glycolysis & Gluconeogenesis |
Glucose, G6P, F1,6BP, 3-PG, PEP, pyruvate, lactate |
Has glycolytic flux increased? Is there a shift toward lactate? |
| TCA Cycle |
Citrate, isocitrate, α-KG, succinate, fumarate, malate |
Are TCA intermediates depleted or accumulated in disease or stress? |
| Oxidative Phosphorylation–Linked |
Redox-related organic acids; NAD⁺/NADH*, FAD/FADH₂* |
Is mitochondrial activity or redox balance altered? |
| Amino Acid–Energy Coupling |
Glutamine, glutamate, BCAAs, alanine, aspartate, others |
Do amino acids feed into the TCA cycle differently between groups? |
| Fatty Acid Oxidation |
Carnitine, short/medium/long-chain acylcarnitines |
Is β-oxidation enhanced or impaired? |
| Ketone Body Metabolism |
β-hydroxybutyrate, acetoacetate, acetone |
Are ketone bodies used as alternative fuels under stress or fasting? |
| High-Energy Compounds |
ATP/ADP/AMP*, creatine, creatinine |
Does energy charge change with treatment or training? |
*Measured and reported where technically feasible for a given matrix and project design.
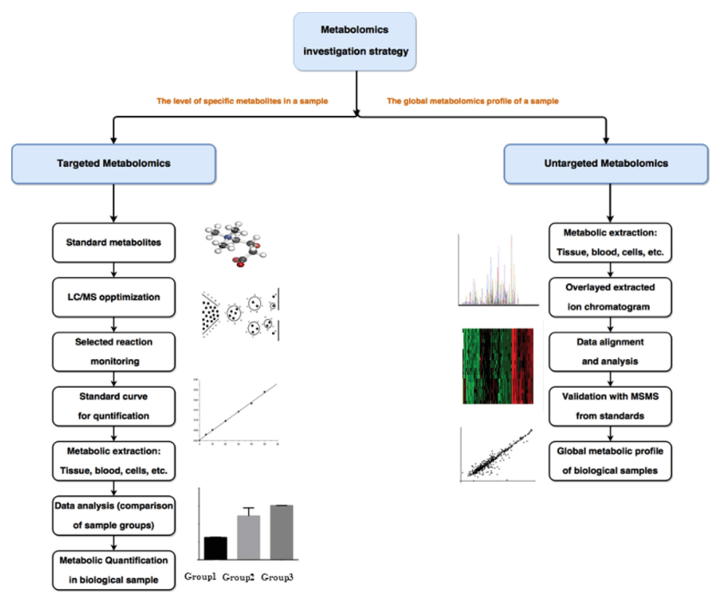
Targeted, Semi-Quantitative, and Exploratory Options
Targeted quantification
- Predefined panel of energy-related metabolites
- Internal standards for improved accuracy
- Best for hypothesis-driven projects
Semi-quantitative profiling
- Expanded list of energy-associated metabolites
- Relative comparison across experimental groups
- Useful for pathway-level interpretation and screening
Untargeted profiling (optional)
- LC–MS-based untargeted metabolomics
- Global metabolite coverage beyond predefined energy panels
- Helps identify additional metabolic processes influenced by your intervention
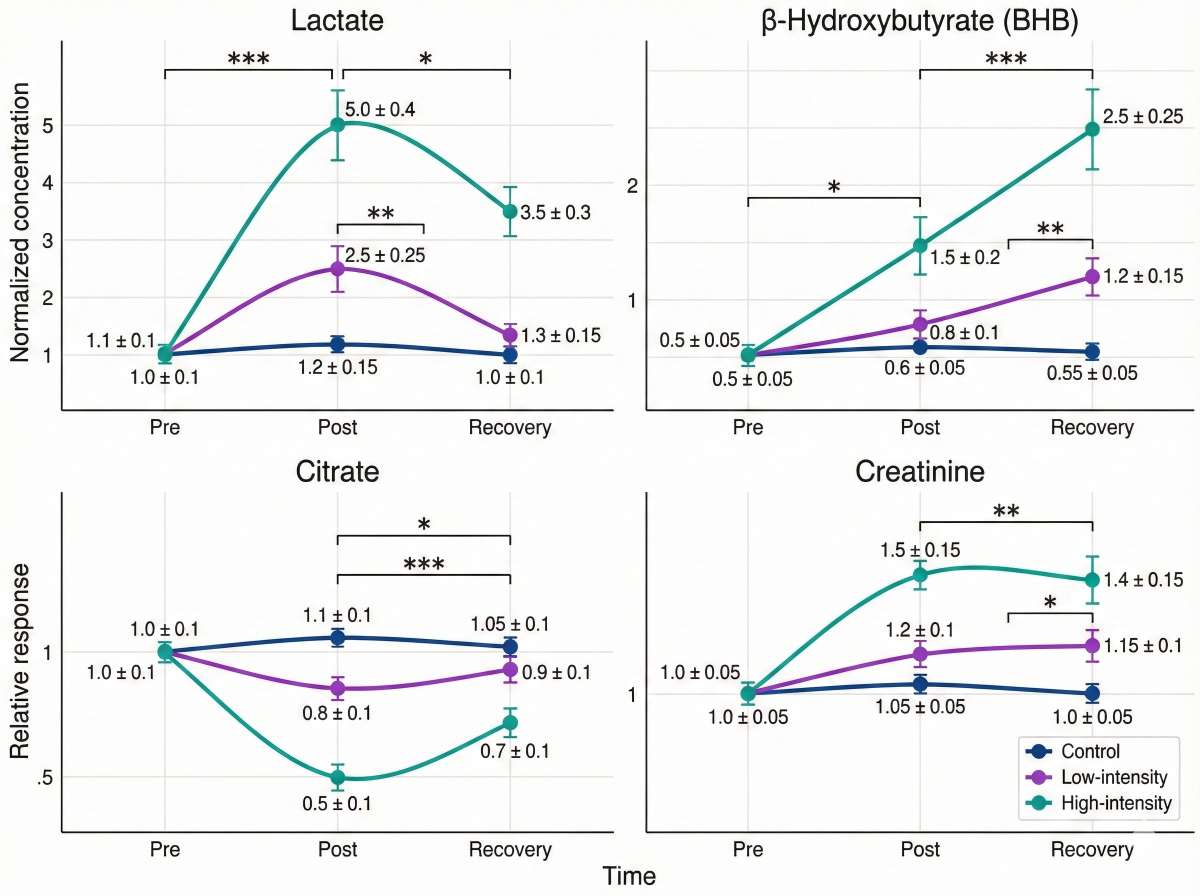
Research Applications of Energy Metabolism Metabolomics
Why Choose Our Energy Metabolism Analysis Services?
- Energy-focused, not generic – Panels and interpretation are built specifically around glycolysis, TCA, fatty acid oxidation, ketone bodies, and amino acid–linked energy pathways.
- High information per run – Up to 100+ energy-related metabolites in a single targeted LC–MS/MS assay, covering central carbon metabolism in one dataset.
- Reproducible, study-ready data – Optimized methods and QC typically keep key markers within low single-digit %CV where feasible, supporting robust group and time-course comparisons.
- Clear, mechanism-oriented outputs – Results are delivered as pathway-level changes (what shifts, how much, and in which direction), making it faster to move from data to experimental decisions.
Analytical Setup and Study Design for Energy Metabolism Profiling
Core LC–MS/MS Platform
- Triple-quadrupole or equivalent LC–MS/MS systems
- Reversed-phase and/or HILIC separation for polar and semi-polar metabolites
- Positive and negative ion modes in scheduled MRM/SRM
- Internal standards and pooled QCs in every batch for robust quantification
Ideal for: focused energy metabolism panels in animal, cell, plant, or microbial studies.
High-Resolution LC–MS for Discovery
- Orbitrap/TOF-class high-resolution LC–MS
- Full-scan acquisition with MS/MS (DDA or DIA)
- Feature detection and annotation for pathway-level exploration
Ideal for: discovery projects and multi-omics integration where you want to see beyond the predefined energy panel.
Stable Isotope Tracing (Optional)
- ¹³C-glucose, ¹³C-glutamine, or other labeled substrates
- Isotopologue-resolved LC–MS/MS methods
- Readouts include labeling patterns in glycolysis, TCA cycle, and related routes
Best suited for: flux-oriented studies that aim to quantify how carbon flows through energy pathways, not just how much metabolite is present.
Key Technical Parameters
| Category |
Typical Specification* |
| Chromatography |
Reversed-phase C18 and/or HILIC, 10–30 min gradients |
| Ionization |
ESI, positive and negative modes |
| Mass analyzers |
Triple quadrupole LC–MS/MS; Orbitrap/TOF for high-resolution runs |
| Mass resolution (HRMS) |
Up to ~30,000–60,000 FWHM at m/z 200 (method-dependent) |
| Quantitative range |
~3–4 orders of magnitude (panel- and matrix-dependent) |
| Typical LOQ |
Low nM to low µM for many energy metabolites in plasma/serum |
| QC strategy |
Internal standards, pooled QC samples, retention time and signal checks |
| Throughput |
Dozens to hundreds of samples per batch, depending on method and design |
Sample Types and Biological Matrices for Energy Metabolism Studies
The platform supports a wide range of matrices. Typical requirements (exact amounts depend on panel and matrix) are shared during project setup.
| Category |
Example Matrices |
Typical Volume / Amount (Guideline) |
| Biofluids |
Plasma, serum, urine, CSF*, saliva*, culture media |
Tens to hundreds of µL |
| Animal tissues |
Liver, muscle, heart, adipose, brain, kidney, tumor |
20–50 mg wet weight (or as discussed) |
| Plant materials |
Leaf, root, stem, seed, fruit |
20–50 mg wet weight (or as discussed) |
| Microbial samples |
Cell pellets, biofilms, fermentation broth |
Pellet from defined OD / volume |
| In vitro models |
Adherent and suspension cells, organoids, spheroids |
Defined cell number or protein content |
*Feasibility depends on sample volume, stability, and expected concentration range.
Collection and Storage Guidance
- Biofluids: clear instructions for fasting, anticoagulants, processing time, and storage
- Tissues & cells: standardized quenching and extraction procedures to preserve labile metabolites
- Storage: typically at −80 °C with minimal freeze–thaw cycles
Data, Reports, and Bioinformatics Deliverables
At project completion, you typically receive:
- Raw data files (e.g., MS data, where applicable)
- Processed data matrix (sample × metabolite, with annotations)
- Statistical analysis results (p-values, fold changes, FDR/adjusted p where applied)
- Pathway analysis outputs focused on energy metabolism
- Graphical figures suitable as a starting point for publication/slide decks
- Summary report explaining methods, major findings, and interpretation
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Macrophage-associated lipin-1 promotes β-oxidation in response to proresolving stimuli
Schilke, R. M., Blackburn, C. M., Rao, S., Krzywanski, D. M., Finck, B. N., & Woolard, M. D.
Journal: Immunohorizons
Year: 2020
DOI: https://doi.org/10.4049/immunohorizons.2000047
Mechanisms underlying neonate-specific metabolic effects of volatile anesthetics
Stokes, J., Freed, A., ... & colleagues
Journal: eLife
Year: 2021
DOI: https://doi.org/10.7554/eLife.65400