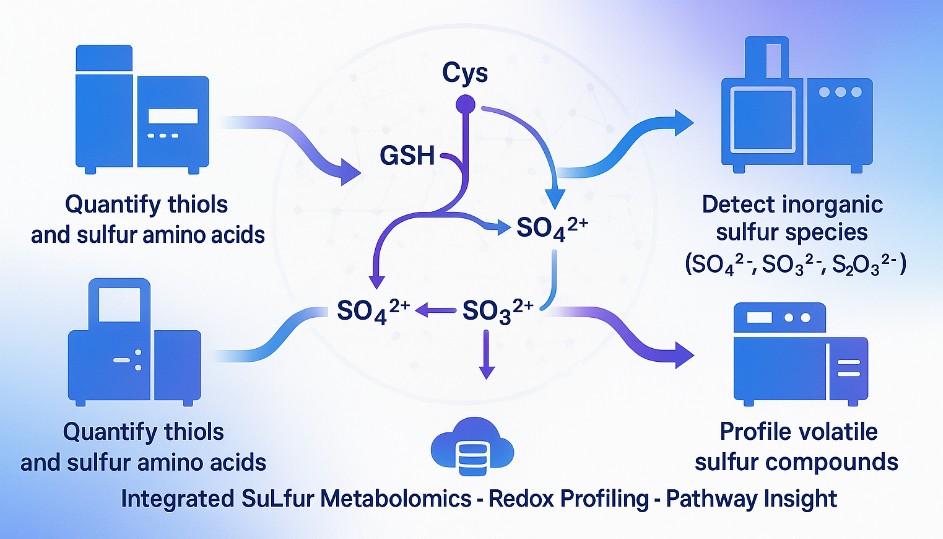
What Is Sulfur Metabolism and Why Analyze Sulfur Compounds?
Sulfur is a central element in both biological systems and industrial processes. In living organisms, it participates in redox regulation, enzyme activation, amino acid synthesis, and structural stabilization through cysteine, methionine, and glutathione pathways. In industrial and bioprocess settings, sulfur compounds influence product yield, stability, and even sensory quality—particularly in fermentation, pharmaceuticals, and food production.
Analyzing sulfur metabolism and sulfur-containing compounds provides essential insights into redox balance, metabolic flux, and product quality control. Through accurate speciation—distinguishing sulfate, sulfite, thiosulfate, and organosulfur species—researchers can connect biochemical pathways with process performance or environmental behavior.
For R&D teams, such analysis supports:
- Monitoring sulfur flux in cell cultures or fermentation systems.
- Identifying unwanted sulfur contaminants or volatile compounds.
- Evaluating antioxidant and redox-active metabolites under stress or formulation changes.
By integrating targeted sulfur compound profiling, Creative Proteomics helps clients translate complex sulfur chemistry into actionable, decision-ready data.
What's Included in Our Sulfur Analysis Service
Creative Proteomics offers targeted sulfur analysis that address the full complexity of sulfur metabolism. Each module is optimized for a specific class of sulfur compounds or analytical objective, allowing clients to choose the combination that best fits their research or quality control needs.
Our service includes the following analytical project types:
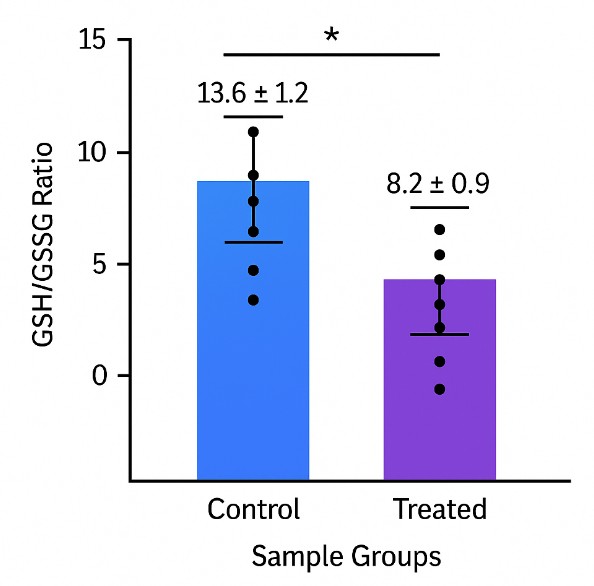
- Redox-focused thiol panels – Tailored to measure reduced and oxidized thiols in parallel, preserving native redox status through stabilizing reagents and derivatization workflows.
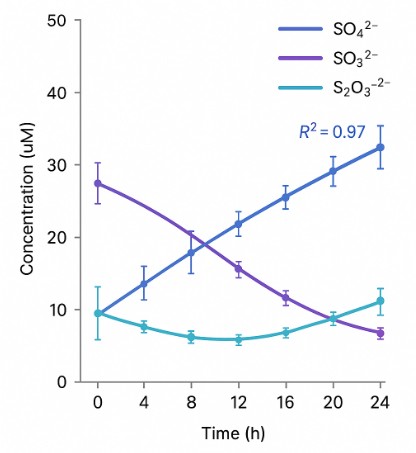
- Inorganic sulfur tracking – Designed for environmental, fermentation, or stress biology studies, this panel quantifies reactive sulfur oxyanions involved in redox cycling.
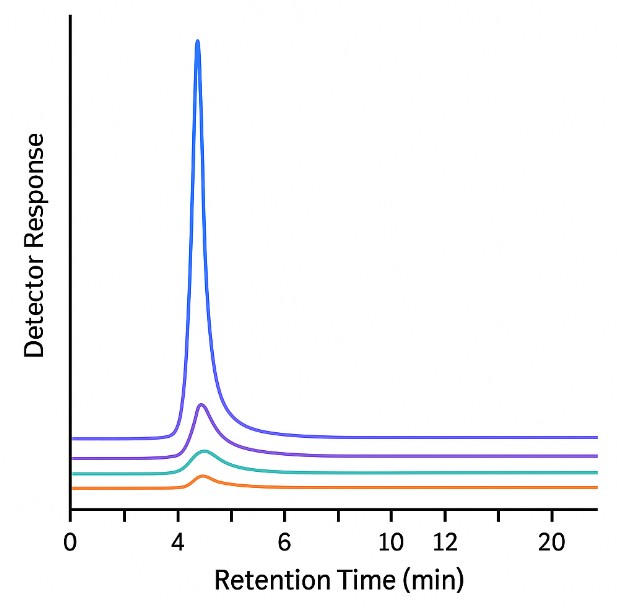
- Volatile sulfur compound profiling – Enables trace detection of sulfur volatiles that influence flavor, safety, or metabolic state, using gas-phase separation and sulfur-selective detection.
- Sulfur amino acid metabolism mapping – Covers key intermediates in methionine and taurine pathways, useful for understanding nutritional status, metabolic flux, or engineered strain behavior.
- Sulfolipid investigation – Supports studies involving sulfur-linked lipids such as SQDG, relevant in microbial lipidomics and stress responses.
- Total sulfur evaluation – Applicable for raw material screening, process monitoring, or formulation consistency checks.
Each of these analysis modules can be requested individually or bundled as part of a customized target panel, depending on sample type, detection limits, and research objectives.
Full Analyte List: Representative Sulfur Compounds We Detect
| Category |
Representative Compounds |
| Inorganic Sulfur Species |
Sulfate (SO42-), Sulfite (SO32-), Thiosulfate (S2O32-), Free sulfide (HS-), Elemental sulfur (S0) |
| Thiols & Disulfides |
Cysteine, Cystine, Glutathione (GSH, GSSG), Homocysteine, Cysteinylglycine (Cys-Gly), N-acetylcysteine (NAC) |
| Oxidized Derivatives |
Cysteic acid, Cysteine sulfinic acid, Methionine sulfoxide, Methionine sulfone |
| Volatile Sulfur Compounds |
Dimethyl sulfide (DMS), Dimethyl disulfide (DMDS), Dimethyl trisulfide (DMTS), Methanethiol (CH3SH), Allyl/Propyl sulfides |
| Sulfur-Containing Amino Acids |
Methionine, Taurine, Hypotaurine, S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH) |
| Sulfur-Linked Lipids |
Sulfoquinovosyldiacylglycerols (SQDGs), Other sulfolipids (upon request) |
| Total Sulfur (Bulk Measures) |
Elemental sulfur (solid/liquid), Total sulfur content (ICP-MS/OES or combustion-IR methods) |
Why Choose Our Sulfur Analysis Service?
- Quantitative Speciation (R2 ≥ 0.995):
All sulfur species are quantified against multi-point standard curves with isotope-labeled internal standards where applicable.
- Thiol Stability Preserved:
On-receipt derivatization (e.g., with NEM, mBBr) prevents thiol oxidation, ensuring accurate GSH/GSSG and Cys/Cystine ratios.
- Matrix-Matched Recovery (85–115%):
Calibration is performed in matched matrices (serum, broth, buffers) to minimize suppression; spike recovery data is reported per batch.
- Dual Volatile + Non-Volatile Coverage:
LC–MS/MS + GC–SCD platforms detect both stable metabolites and low-ppb volatiles like DMS and methanethiol.
- Low CV% Across Replicates:
Method CVs typically ≤8% for most species in clean matrices; precision data is included with results.
Main Techniques for Sulfur and Sulfur Compound Quantification
Our sulfur compound analysis service is built on three core analytical platforms, each selected based on compound class, matrix complexity, and required sensitivity. These methods support accurate quantification of both inorganic and organosulfur species, including redox-active thiols and trace-level volatiles.
Main Analytical Platforms
| Method |
Instruments |
Typical Applications |
Key Parameters |
| LC–MS/MS |
Agilent 6495C |
Thiols (Cys, GSH, GSSG), sulfur amino acids, oxidized derivatives |
Derivatization (e.g., NEM, mBBr), isotope-dilution MRM, LOQ ~ nM |
| IC–ICP-MS |
Thermo ICS-6000 + iCAP RQ ICP-MS |
Inorganic sulfur species (SO42-, SO32-, S2O32-, HS-) |
Gradient elution, element-specific detection, R2 ≥ 0.995 |
| GC–MS / GC–SCD |
Agilent 7890/5977B + Sulfur Chemiluminescence Detector |
Volatile sulfur compounds (DMS, methanethiol, DMDS) |
Headspace-SPME, sulfur-specific detector, sub-ppb sensitivity |
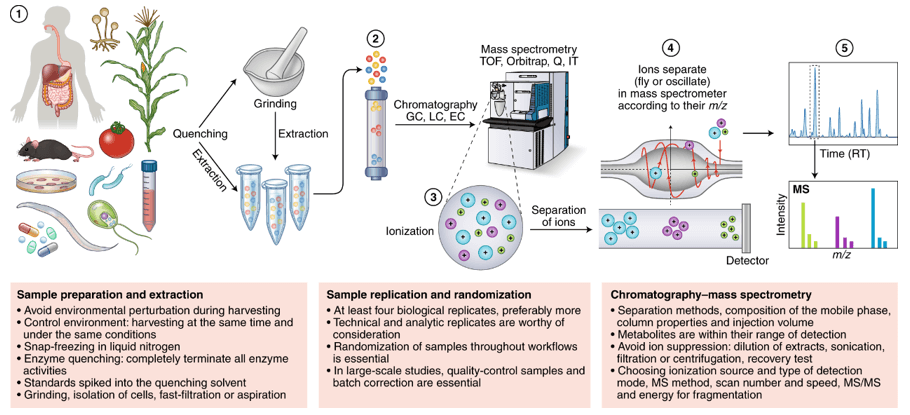
Sulfur Compound Analysis Workflow: From Sample Prep to Data Delivery
How to Prepare and Submit Samples for Sulfur and Sulfur Compound Analysis
| Matrix Type |
Minimum Volume / Mass |
Recommended Container |
Preservative or Treatment |
Storage & Shipping |
Notes |
| Cell lysate / tissue |
50–100 mg or ≥200 µL |
Low-bind tube (1.5 mL) |
Add thiol stabilizer (e.g., NEM or IAA) |
Cold chain, protect from light |
Avoid repeated freeze–thaw cycles |
| Culture supernatant / media |
≥500 µL |
Polypropylene vial, tightly capped |
Optional thiol stabilizer |
Cold chain transport |
Record additives and pH |
| Serum / plasma |
≥200 µL |
EDTA or heparin tube, aliquoted |
Add stabilizer if thiol analysis required |
Keep cold during transit |
Avoid hemolysis |
| Fermentation broth |
≥5 mL |
Sterile sampling bottle |
Mix gently before transfer |
Ship refrigerated |
Note fermentation stage |
| Powder / solid material |
≥1 g |
Clean sealed jar or glass vial |
None required |
Ambient or refrigerated (as suitable) |
Provide SDS or composition sheet |
| Food / beverage sample |
≥100 mL or 200 g |
Food-grade bottle |
None required |
Cold chain recommended |
Note carbonation/alcohol content |
| Buffers / excipients |
≥5 mL |
Compatible chemical-resistant vial |
None required |
Cold chain shipping |
Provide formulation details |
Deliverables: What You Receive from Sulfur Compound Analysis
- Analytical Report (PDF): Compound list, methods, calibration, and QC summary.
- Processed Data (Excel/CSV): Concentrations, LOD/LOQ, recoveries, and CV%.
- Raw Files: LC–MS/MS, IC–ICP-MS, GC–MS data with metadata and logs.
- Chromatograms & Spectra: Annotated peaks, calibration plots, and overlays.
- Visual Summaries: Redox ratios, speciation charts, and time-course trends.
- QC Appendix: Linearity (R2), recovery %, blanks, and stability metrics.
- Optional Add-ons: Custom dashboards or sulfur pathway mapping.
Where Amino and Nucleotide Sugar Analysis Makes an Impact
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566