Gluconeogenesis Analysis Service
Submit Your Inquiry- Service Details
- Demo
- FAQ
- Publications
What is Gluconeogenesis?
Gluconeogenesis is a central metabolic pathway in which glucose is synthesized de novo from non-carbohydrate substrates such as lactate, glycerol, and glucogenic amino acids. This process is crucial for maintaining blood glucose levels during prolonged fasting, intensive exercise, or carbohydrate-restricted conditions. Predominantly occurring in the liver and kidneys, gluconeogenesis is tightly regulated and interconnected with other key metabolic pathways, including glycolysis, the tricarboxylic acid (TCA) cycle, and amino acid metabolism.
Creative Proteomics's gluconeogenesis analysis service is designed to deliver high-resolution, quantifiable, and reproducible insights into this complex pathway. Utilizing state-of-the-art analytical platforms and bioinformatics, we enable researchers to investigate the flux, regulation, and interaction of gluconeogenic intermediates and enzymes under various physiological and experimental conditions.
Advantages of Gluconeogenesis Analysis Service
- Quantitative Accuracy: Our isotope-labeled tracing methods provide quantitation with CVs <10% across biological replicates, enabling precise flux measurements.
- High Throughput Capability: Analyze up to 200 samples per run with batch processing to meet high-volume project needs.
- Wide Dynamic Range: Detection range of 10 nM to 100 µM, ensuring both trace and abundant metabolites are captured effectively.
- Pathway-Integrated Interpretation: Integrated metabolic modeling and pathway visualization support downstream interpretation and hypothesis generation.
- Time-Resolved Kinetics: Kinetic analysis with time-course sampling from 5 min to 24 h post-substrate administration.
- Cross-Species Compatibility: Validated in human, mouse, rat, and other model organisms for translational research.
Gluconeogenesis Analysis Service Offered by Creative Proteomics
- Stable isotope-resolved metabolomics using 13C/15N tracers
- Quantitative flux analysis through the gluconeogenesis pathway
- Targeted quantification of key gluconeogenesis intermediates via LC-MS/MS and GC-MS
- Enzyme activity profiling for PEPCK, FBPase, G6Pase, and related enzymes
- Time-course sampling and dynamic metabolite profiling
- Comprehensive mapping of gluconeogenesis and related metabolic pathways
- Visualization of metabolic shifts through custom pathway diagrams
- Customized statistical analysis and biological interpretation
- Multi-omics data integration (metabolomics + transcriptomics/proteomics)
- Species-specific method customization and tracer adaptation
List of Detected Gluconeogenesis and Related Metabolites
Core Gluconeogenesis Intermediates
| Metabolite | Role in Pathway |
|---|---|
| Glucose-6-phosphate | Final step before glucose export |
| Fructose-6-phosphate | Key reversible intermediate |
| Fructose-1,6-bisphosphate | Rate-limiting step (FBPase target) |
| Glyceraldehyde-3-phosphate | Triose-phosphate isomerization step |
| Dihydroxyacetone phosphate (DHAP) | Precursor from glycerol metabolism |
| 1,3-Bisphosphoglycerate | Energy-coupled conversion step |
| 3-Phosphoglycerate | Downstream product of PEP |
| 2-Phosphoglycerate | Intermediate before PEP conversion |
| Phosphoenolpyruvate (PEP) | Critical entry point to gluconeogenesis |
| Pyruvate | Central link to TCA and amino acids |
| Oxaloacetate | Mitochondrial substrate for PEP |
Connected TCA Cycle Intermediates
| Metabolite | Role in Metabolism |
|---|---|
| Malate | Shuttle for oxaloacetate |
| Fumarate | Intermediate linking to amino acids |
| α-Ketoglutarate | Links amino acids and TCA cycle |
| Succinyl-CoA | Entry from odd-chain fatty acids |
| Citrate | Indicator of acetyl-CoA levels |
| Isocitrate | TCA progression step |
Amino Acid–Derived Gluconeogenic Substrates
| Amino Acid | Metabolic Relevance |
|---|---|
| Alanine | Transaminated to pyruvate |
| Glutamine | Enters via α-ketoglutarate |
| Glutamate | Key nitrogen donor in transamination |
| Aspartate | Converted to oxaloacetate |
Lipid and Glycerol Pathway Intermediates
| Metabolite | Pathway Link |
|---|---|
| Glycerol | Major substrate during fasting |
| Glycerol-3-phosphate | Intermediate via glycerol kinase |
| Acetyl-CoA | Regulates pyruvate carboxylase |
Energy and Redox State Indicators
| Metabolite | Physiological Role |
|---|---|
| NADH / NAD⁺ | Affects flux directionality |
| ATP / ADP | Energy balance for enzyme activation |
| Urea | Linked to amino acid catabolism |
| Phosphocreatine | Energy buffer, indirect flux marker |
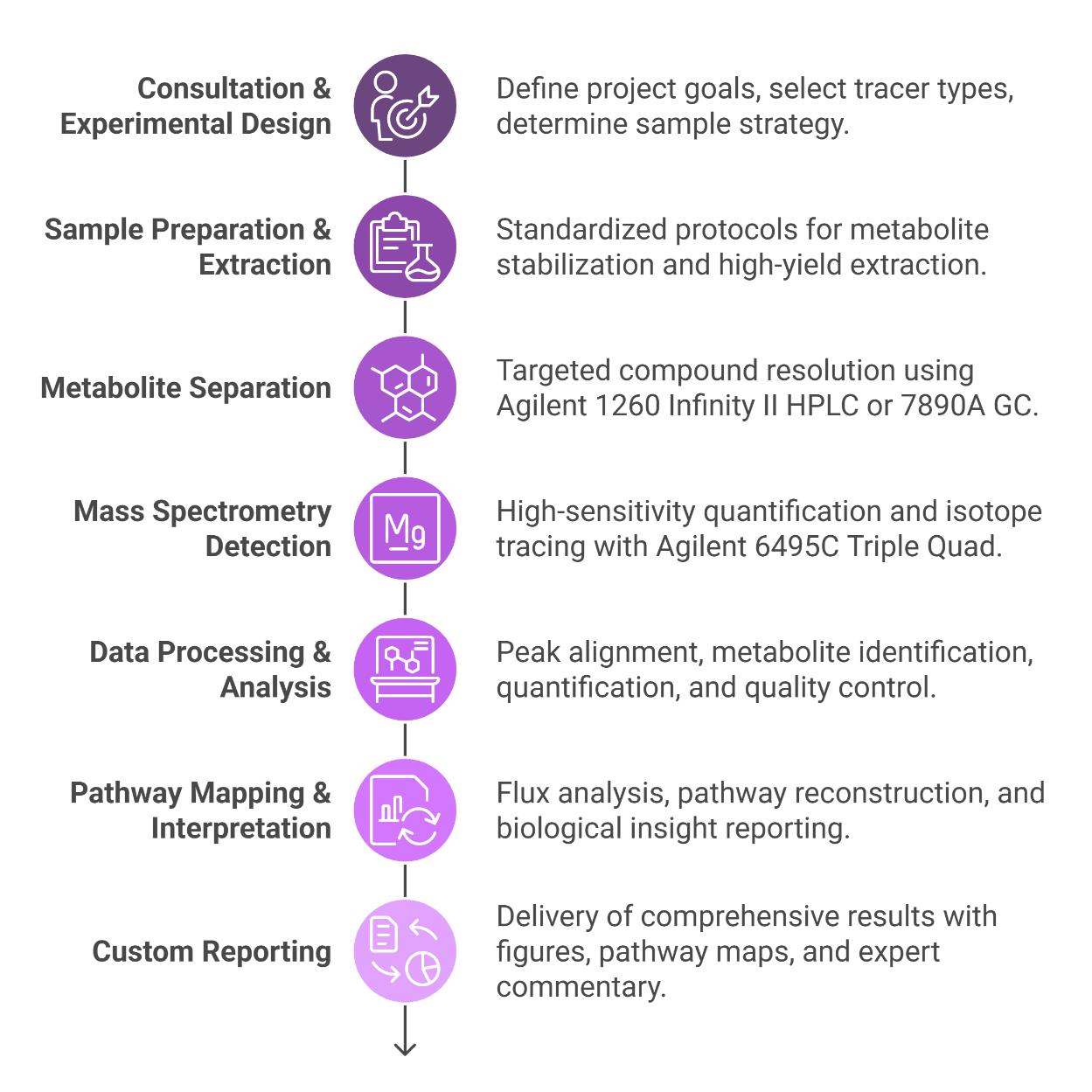
Workflow for Gluconeogenesis Analysis Service
Technology Platform for Gluconeogenesis Analysis Service



Agilent 6495C Triple quadrupole (Figure from Agilent)
Agilent 7890B-5977A (Figure from Agilent)
Agilent 1260 Infinity II HPLC (Fig from Agilent)
Sample Requirements for Gluconeogenesis Analysis Service
| Sample Type | Required Volume / Mass | Notes |
|---|---|---|
| Cell Pellet | ≥ 1 × 10⁷ cells | Washed with cold PBS; snap-frozen |
| Adherent Cells | Equivalent to 1 × 10⁷ cells | Preferably cultured in 6-well or 10 cm dishes |
| Tissue (e.g., liver, kidney) | ≥ 50 mg (wet weight) | Flash-frozen in liquid nitrogen immediately |
| Serum / Plasma | ≥ 100 µL | Collected in EDTA or heparin tube |
| Urine | ≥ 500 µL | First morning sample preferred |
| Culture Medium | ≥ 500 µL | Without phenol red or serum (for isotope studies) |
| Bacterial / Yeast Pellet | ≥ 100 mg (wet weight) | Harvested during mid-log phase, freeze-dried optional |
| CSF (Cerebrospinal Fluid) | ≥ 50 µL | For specific brain glucose metabolism studies |
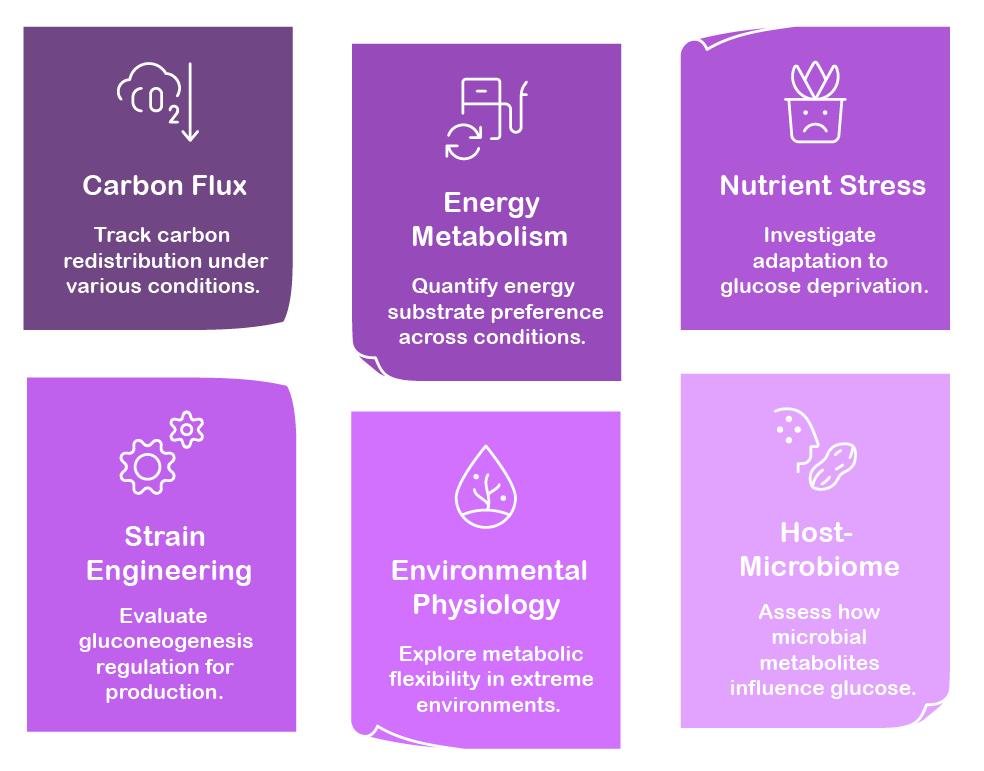
Applications of Gluconeogenesis Assay Service
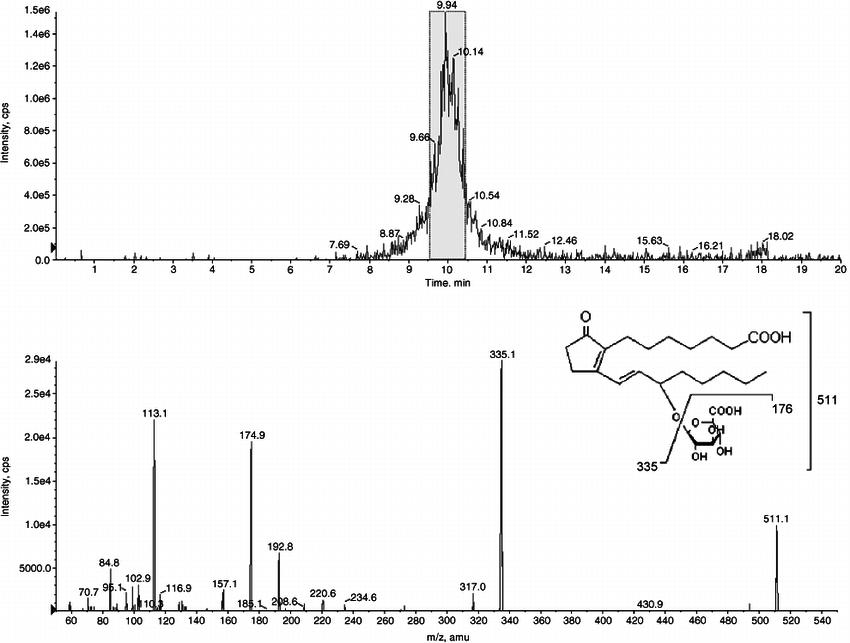 LC-MS/MS analysis of the PGB 1 glucuronide biosynthesized by HL microsomes (Little, Joanna M., et al., 2004).
LC-MS/MS analysis of the PGB 1 glucuronide biosynthesized by HL microsomes (Little, Joanna M., et al., 2004).
Reference
- Little, Joanna M., et al. "Glucuronidation of oxidized fatty acids and prostaglandins B1 and E2 by human hepatic and recombinant UDP-glucuronosyltransferases." Journal of lipid research 45.9 (2004): 1694-1703. https://doi.org/10.1194/jlr.M400103-JLR200
Q1: Can I analyze both intracellular and extracellular metabolites in one experiment?
A1: Yes. We offer separate or combined profiling of intracellular and extracellular fractions, depending on your study design. Please specify during consultation.
Q2: Do you support isotope-labeled tracer optimization for custom substrates?
A2: Yes. We provide consultation on tracer selection and labeling strategies, including custom tracers like [U-13C] glycerol or [2-13C] pyruvate.
Q3: What normalization methods are used for metabolite quantification?
A3: We offer normalization by cell number, protein content, tissue weight, or external/internal standards, based on your study needs.
Q4: Can I submit samples in organic solvents (e.g., methanol extract)?
A4: Yes. We accept both raw biological samples and pre-extracted metabolites. Please contact us for solvent compatibility and volume requirements.
Q5: What if I only have a limited sample amount?
A5: We can work with minimal sample input and offer pre-analysis consultation to adjust detection strategies for low-volume samples.
Q6: Do you provide flux calculation using isotope labeling data?
A6: Yes. We offer metabolic flux analysis (MFA) including isotopomer distribution models and pathway-specific flux quantification.
Q7: Can you help design my gluconeogenesis experiment from scratch?
A7: Absolutely. We provide end-to-end experimental design support, including controls, sampling time points, and data interpretation strategy.
Q8: Is batch effect correction included in your data analysis?
A8: Yes. Our pipeline includes statistical correction for batch effects, ensuring high comparability across sample groups.
Q9: Will I receive pathway visualization in the report?
A9: Yes. Our standard report includes annotated pathway maps, flux diagrams (if applicable), and detailed metabolite heatmaps.
Q10: Can you perform analysis on archived or long-stored samples?
A10: Yes, provided that the samples have been stored properly at -80°C without multiple freeze-thaw cycles. We can help evaluate sample integrity.
Q11: What is the detection limit of the Agilent 6495C Triple Quadrupole for gluconeogenic intermediates?
A11: The Agilent 6495C LC/MS system achieves a detection limit of 0.1 fmol/μL for key metabolites like phosphoenolpyruvate (PEP) and oxaloacetate, with a linear dynamic range spanning six orders of magnitude (R² > 0.998).
Q12: How does the Agilent 7890A GC ensure reproducibility for glycerol quantification?
A12: Using the VF-WAXms column (30 m × 0.25 mm), we achieve a retention time CV <0.5% for glycerol in plasma samples. The system's electron impact ionization (EI) mode ensures 99% isotopic purity for [2H]-labeled tracers
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal: Transplantation and Cellular Therapy
Year: 2024
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024