Integrative Metabolome and Proteome Analysis
Submit Your Inquiry
Proteins are the performers of the functions of life activities. Proteomics studies the composition and activity patterns of intracellular proteins and protein-protein interactions at a holistic level. Metabolomics explores key scientific questions related to cellular metabolism in living organisms by qualitatively describing and quantitatively characterizing changes in small molecule metabolites in different biological matrices. Metabolomics serves not only as a depictor of phenotypic states, but also as a functionally regulated active metabolite that can modulate protein-protein interactions, alter enzyme activities, lead to changes in protein stability, and thus feed back to regulate the metabolic state of the organism.
Integrated proteomics and metabolomics analysis refers to the normalization and statistical analysis of bulk data from the proteome and metabolome. Combined with metabolic pathway enrichment and correlation analysis, molecular models of downstream metabolite changes and metabolic enzyme regulation mechanisms are constructed, thus providing a data base for subsequent in-depth experiments and analysis. The combined proteomic and metabolomic analysis can systematically depict the metabolic regulatory network of biological organisms.
Creative Proteomics has established a platform for proteomic and metabolomic analysis, which allows the screening of key metabolic enzymes (proteins) and metabolites and metabolic pathways by comparing the differences in protein or metabolite expression. At the same time, the results of the analysis of the two groups can also be cross-validated, which helps to study the regulatory mechanisms of physiological and pathological processes more systematically and comprehensively, and to discover new biomarkers, etc.
Technology Roadmap for
Integrative Metabolome and Proteome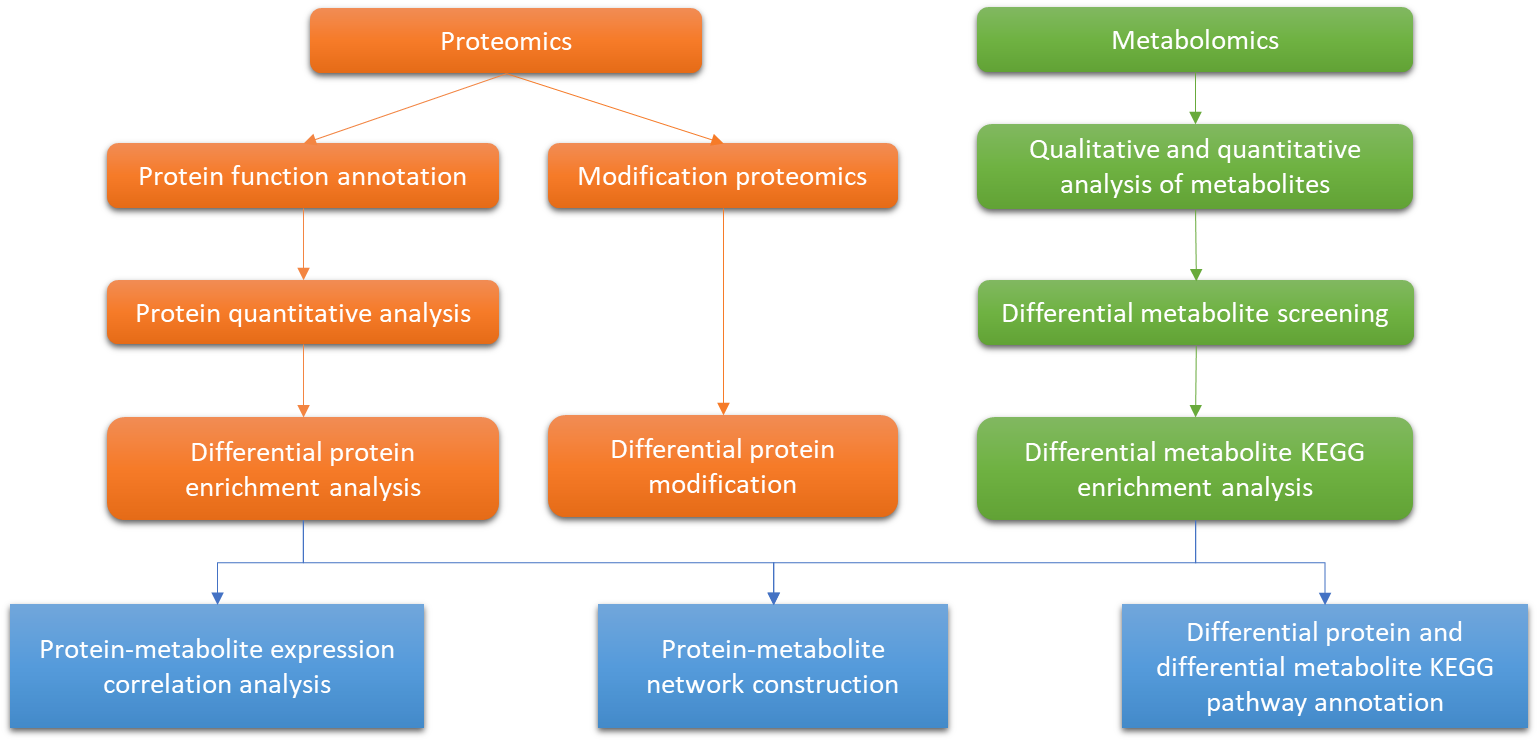
Sample Submission
| Sample Type | Proteomics | Metabolomics |
| Plant tissue | Fresh weight ≥1g | 3g recommended, 1g minimum |
| Animal tissue | ≥200mg | 200mg recommended, minimum 50mg |
| Plasma/Serum | ≥20 uL | ≥20 uL |
| Cell | ≥5×106 or cell precipitation not less than 50μL | ≥5×106 or cell precipitation of not less than 50μL |
Data Analysis for
Integrative Metabolome and Proteome| Analysis Type | Analyze Content |
| KEGG metabolic pathway analysis | KEGG pathway visualization, comparative pathway analysis, enrichment analysis |
| Correlation analysis | Correlation coefficient matrix heat map. Correlation analysis hierarchical clustering heat map. Correlation coefficient regulatory network analysis |
| PCA comparative analysis | Comparative proteomic and metabolomic PCA analysis |
| Multivariate statistical integration analysis | O2PLS correlation analysis. Screening of OPLS correlation variables. |
| WGCNA analysis | Weighted gene co-expression network analysis |
If you want to know more, please contact us. Looking forward to cooperating with you.
For Research Use Only. Not for use in diagnostic procedures.