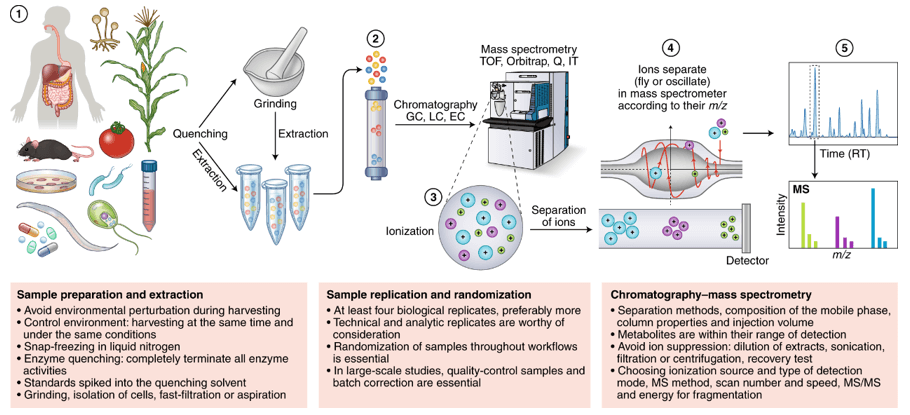
Metabolic pathways analysis involves the systematic study and mapping of the biochemical processes that occur within an organism. This analysis provides crucial insights into the flow of metabolites, the regulation of various biochemical reactions, and the interplay of different pathways. The primary objectives of Metabolic Pathways Analysis include:
- Identifying Key Metabolites: Understanding the metabolites involved in various pathways is vital for studying diseases, drug metabolism, and cellular responses.
- Mapping the Flow of Metabolites: It helps in tracking the movement of metabolites within different cellular compartments and pathways.
- Elucidating Regulatory Mechanisms: Discovering the control mechanisms that govern the activity of specific enzymes and metabolic reactions.
- Biological Insights: Offering a comprehensive view of the metabolic state of an organism, which can be instrumental in research, diagnostics, and therapeutic development.
Metabolic pathways are vast networks of biochemical reactions that maintain biological function and include highly conserved metabolic pathways composed of a series of chemical reactions necessary to maintain and regulate life activities. Metabolic pathway analysis can identify metabolite clusters associated with key cellular signaling and metabolic networks and provide insight into the biological potential of differentially expressed metabolites. In metabolic pathways, there may be many metabolites with very similar chemical structures and polarities, and multiple pathways are needed to obtain quantitative information on multiple species of metabolites.
Creative Proteomics provides metabolic pathway analysis solutions for a wide range of biological samples such as blood, urine, saliva, tears, animal and plant tissues, providing metabolite assays covering multiple metabolic pathways such as amino acid, tricarboxylic acid cycle, glycolysis, nucleic acid metabolism, and hormone metabolism.
Creative Proteomics' Metabolic Pathways Analysis Services
At Creative Proteomics, we offer a range of specialized services for metabolic pathways analysis, each tailored to meet the unique needs of our clients. Our offerings include:
Metabolite Profiling
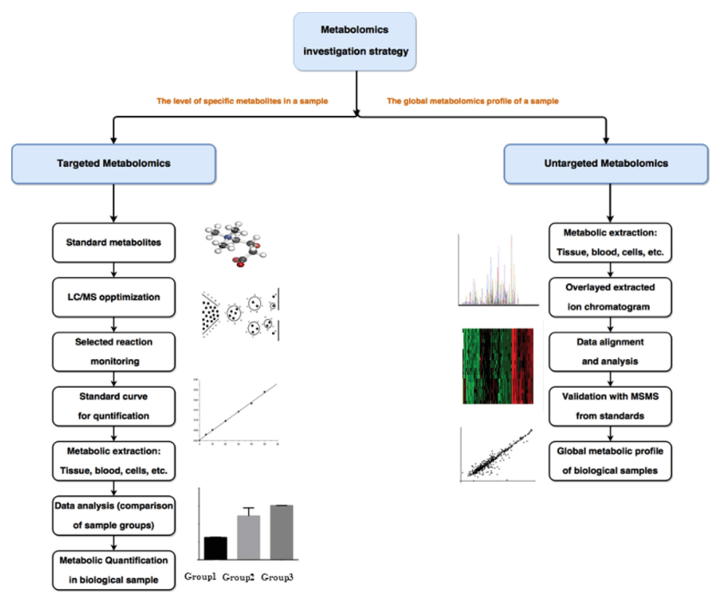
Metabolites are the small molecules that participate in various metabolic reactions within biological systems. Our metabolite profiling services aim to identify, quantify, and characterize these metabolites. Creative Proteomics offers two main approaches in this domain:
- Targeted Metabolomics: This approach focuses on the analysis of specific metabolites of interest. It is particularly valuable when you have a predefined set of metabolites they want to investigate. Targeted Metabolomics allows for high sensitivity and precision.
- Untargeted Metabolomics: In contrast, untargeted metabolomics provides a comprehensive analysis of all detectable metabolites in a given sample. It is an excellent choice for gaining a holistic understanding of the metabolic landscape within a biological system.
Enzyme Activity Assays
Enzymes are the catalysts that drive biochemical reactions within metabolic pathways. Creative Proteomics offers the following enzyme activity assays:
- Enzyme Kinetics Analysis: This service involves quantifying enzyme activity and characterizing their kinetics. It provides insights into reaction rates, substrate specificity, and the regulatory mechanisms governing enzyme function.
- Pathway-Specific Enzyme Assays: Focused assessments of enzymes within a particular pathway.
Flux Analysis
- Stable Isotope Tracing: This method involves introducing stable isotope-labeled substrates into the biological system and tracking their fate. It enables the determination of metabolic flux rates and can reveal the routes metabolites take within the pathways.
- Flux Balance Analysis: This computational approach models and simulates metabolic flux within a cellular system. It can predict the impact of substrate changes, evaluate the effects of genetic perturbations, optimize biotechnological processes, and assess metabolic network efficiency under various scenarios.
Metabolic Pathway Mapping
- Pathway Enrichment Analysis: Identifying enriched pathways within high-throughput omics data.
- Visualization and Interpretation: Clear visualization and interpretation of metabolic pathways.
Metabolic Pathways Analysis Analytical Techniques
| Analytical Technique |
Instrument Models |
Applicability |
Typical Applications |
| Liquid Chromatography-Mass Spectrometry (LC-MS/MS) |
- Thermo Scientific Q Exactive Plus
- Waters Xevo TQ-S |
High-resolution analysis of a wide range of metabolites. |
- Metabolite profiling
- Biomarker discovery
- Drug metabolism studies |
| Gas Chromatography-Mass Spectrometry (GC-MS) |
- Agilent 7890B GC
- Agilent 5977A MSD |
Ideal for volatile metabolites. |
- Analysis of small, volatile compounds
- Environmental monitoring
- Chemical analysis |
| Nuclear Magnetic Resonance (NMR) Spectroscopy |
- Bruker Avance III 600 MHz
- Varian NMR Systems |
Non-destructive, structural elucidation of metabolites. |
- Structure determination of organic compounds
- Metabolite identification
- Protein-ligand interactions |
| Ultra-High Performance Liquid Chromatography (UHPLC) |
- Agilent 1290 Infinity II
- Waters ACQUITY UPLC |
High-throughput analysis of complex samples. |
- Quantitative analysis of various metabolites
- Separation and detection of biomolecules
- Pharmaceutical analysis |
List of Metabolic Pathways Analysis We Can Analyze
Feature metabolic pathways:
If you want to know more, please contact us. Looking forward to cooperating with you.
Reference
- Lu, Z., He, X., et al. (2018). Serum metabolomics study of nutrient metabolic variations in chronic heat-stressed broilers. British Journal of Nutrition, 119(7), 771-781.
Case: Mitochondrial Serine Metabolism and SHMT2 in Breast Cancer Metastasis
Background
The study focuses on understanding the role of serine metabolism, particularly the enzyme SHMT2, in the context of breast cancer metastasis. Metastasis is a complex process involving the selection of cancer cell subpopulations with enhanced fitness and adaptability, leading to treatment resistance and cancer progression. While previous research has explored various molecular processes contributing to metastasis, the impact of metabolic pathway alterations remains an underexplored area.
Samples: The research primarily utilizes the human triple-negative breast cancer cell line, MDA-MB-231, and its metastatic subpopulations (831-BrM and 1833-BoM) along with the parental cell line (231-Parental). The cell lines represent different stages of metastatic progression, making them suitable for investigating metabolic changes associated with metastasis.
Techniques and Methods
Metabolomic Profiling: Metabolomic profiling was conducted on the selected cell lines to identify alterations in metabolic pathways during metastasis. This analysis involved the measurement of various metabolites to identify differences in metabolic profiles.
Gene Expression Analysis: The study examined the expression of key enzymes involved in serine and 1C unit pathways, such as SHMT2, MTHFD2, MTHFD1L, and SHMT1, to understand their role in metastatic cells.
Stable Isotope Tracing: The researchers used [2,3,3-2H]serine tracing to investigate 1C unit pathway flux to glycine and purine nucleotides. This technique allowed them to assess the metabolic activity of the mitochondrial serine and 1C unit pathway.
Genetic Knockdown: The study employed small hairpin RNA (shRNA) to knock down the expression of the oncogenic transcription factor c-Myc, SHMT2, and MTHFD2 to examine their impact on cell proliferation.
In Vivo Models: To assess the relevance of SHMT2 in vivo, the research involved two different models: primary tumor growth in mammary fat pads and breast cancer lung metastasis through tail-vein injection. The growth of tumors was monitored using bioluminescence imaging (BLI).
Results
- Metastatic breast cancer cells exhibited altered metabolic profiles, including changes in glycolysis, the tricarboxylic acid (TCA) cycle, and purine metabolism.
- Elevated expression of c-Myc was observed in metastatic subclones, contributing to their increased proliferation rates.
- Metastatic cells displayed increased mitochondrial serine and 1C unit pathway activity, promoting de novo purine synthesis, essential for their rapid growth.
- Knockdown of SHMT2 impaired primary and metastatic growth in vivo, indicating its importance in breast cancer progression.
- Gene expression analysis and clinical data from human breast cancer patients demonstrated that mitochondrial 1C unit pathway enzyme expression correlated with more aggressive metastatic disease, making them potential prognostic markers.
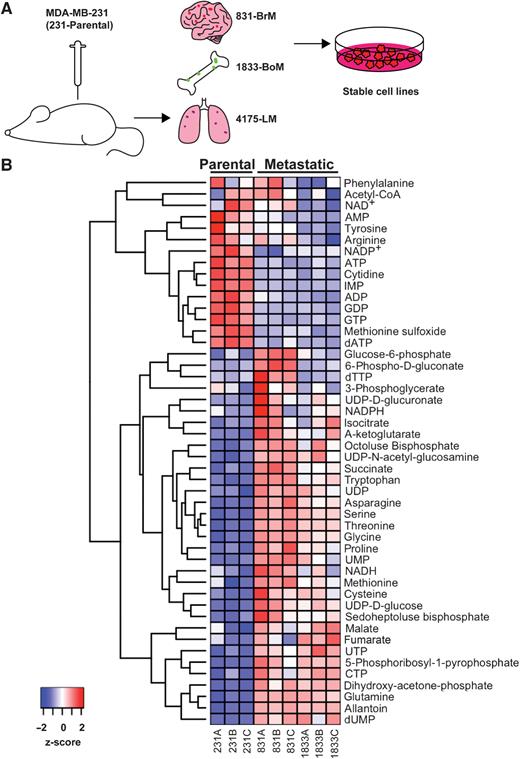 Metastatic breast cancer subclones display an altered metabolic profile.
Metastatic breast cancer subclones display an altered metabolic profile.
Reference
- Li, Albert M., et al. "Metabolic profiling reveals a dependency of human metastatic breast cancer on mitochondrial serine and one-carbon unit metabolism." Molecular Cancer Research 18.4 (2022): 599-611.

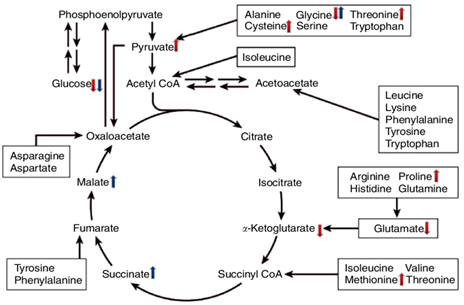
 Metastatic breast cancer subclones display an altered metabolic profile.
Metastatic breast cancer subclones display an altered metabolic profile.