Overview
Plasma, serum or urine are used in most metabolomics studies because these body fluids are readily available and can be collected in a minimally invasive or non-invasive manner. However, metabolic changes in disease are usually tissue-based. To characterize metabolic changes under disease phenotypes, it is beneficial to directly analyze metabolomic changes in animal model tissue samples or human biopsy samples. Tissue collection is invasive, but comparative analysis of metabolite profiles in diseased and non-diseased tissue regions can be performed thereby screening for differential metabolites and facilitating disease mechanism studies.
Metabolomics studies using tissue samples face many challenges such as tissue inhomogeneity, tissue collection, metabolic bursts, tissue homogenization and metabolite extraction. The same sample pretreatment, solvents used, and different extraction protocols can lead to biased results. The choice of targeted or untargeted metabolomics strategy can also influence the metabolic profile analyzed and the biological interpretation of the data obtained. No single extraction method can satisfy that all metabolites can be well extracted. Therefore, the most appropriate compromise extraction method for metabolomic analysis is chosen based on multiple factors such as research needs, throughput, and processing efficiency.
Sometimes we may have limited sample size, such as the limited amount of tissue available in human biopsies and the small amount of tissue available in commonly used mouse models, making it necessary to develop an efficient and tissue-saving metabolite extraction process both for ethical and practical needs.
Creative Proteomics offers customized metabolomics analysis of tissue samples based on the LC-MS/MS platform. Depending on the sample and research needs, we have optimized a variety of sample pre-processing and extraction solutions to simultaneously extract and analyze metabolite related products from a single small amount of tissue sample. We can provide not only absolute quantitative analysis of targeted metabolites or one class of metabolites, but also non-targeted metabolomics analysis of all metabolites to screen for differential metabolites.
What We Can Provide:
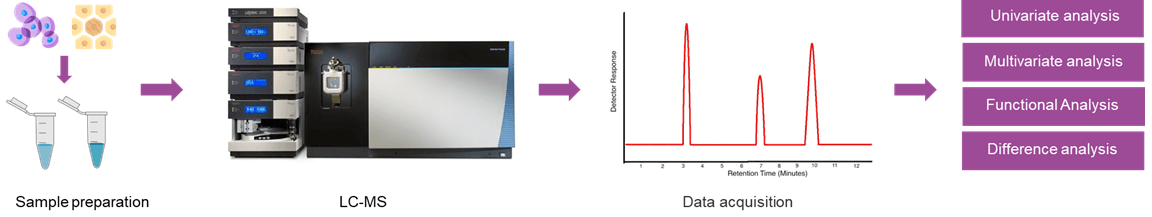
Untargeted metabolomics analysis:
- Metabolic profile analysis
- Screening for differential metabolites
- Metabolic pathway analysis of differential metabolites
- Discover new biomarkers
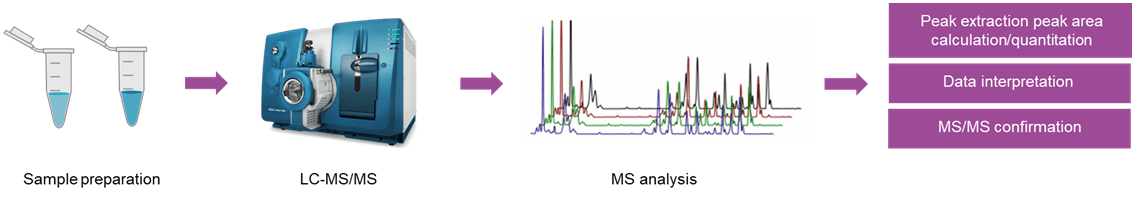
Targeted metabolomics analysis:
- Mode: SRM / MRM
- Analysis content: Standard curve creation. Raw data preprocessing. Absolute quantification of metabolites.
Deliverables
- Experimental procedure
- Parameters of liquid chromatography and MS
- MS raw data files and MS data quality checks
- Bioinformatics analysis report
Turnaround time: 2-6 weeks
Bioinformatics Analysis
| Standard analysis | 1. Data quality control (QC correlation analysis, PCA analysis of total samples) |
| 2. Qualitative and quantitative results of metabolites |
| 3. Differential metabolite screening (PCA, PLS-DA dimensionality reduction treatment, volcano map) |
| 4. Differential metabolite analysis: Venn diagram, cluster heat map, metabolite correlation analysis, Z-score analysis, pathway analysis, ROC analysis |
| Personalized analysis | Weighted gene co-expression network analysis (WGCNA) |
| One way ANOVA |
Creative Proteomics provides metabolomics services for tissue samples, and will deliver accurate and detailed data and analysis reports to you. If you want to detect other metabolites but have not found them, you can tell us through the inquiry form, and our technicians will communicate with you.
For Research Use Only. Not for use in diagnostic procedures.