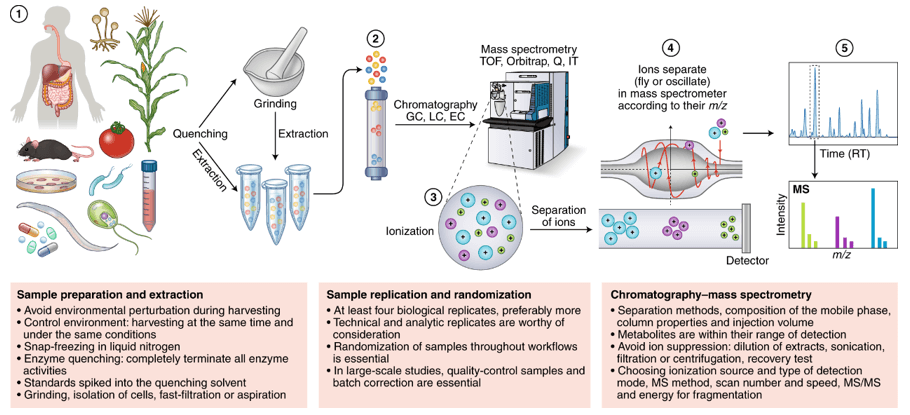
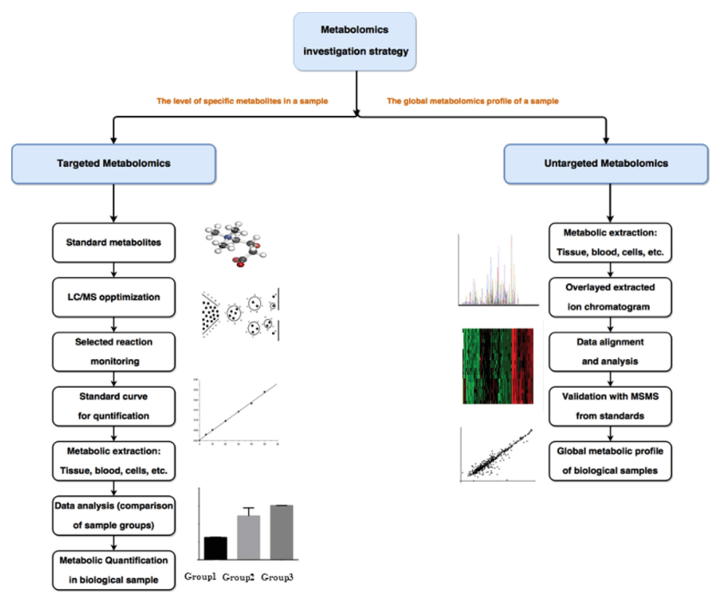
Fecal metabolomics is a new tool to explore the links between microbial composition, host phenotype and complex genetic traits. Creative Proteomics has built a metabolomic analysis platform for feces samples using high-resolution mass spectrometry. We will use the appropriate sample pretreatment protocol for your experimental purposes, significantly improving the accuracy of feces metabolite identification and quantification of metabolites.
What We Can Provide:
Untargeted metabolomics analysis:
- Metabolic profile analysis
- Screening for differential metabolites
- Metabolic pathway analysis of differential metabolites
- Discover new biomarkers
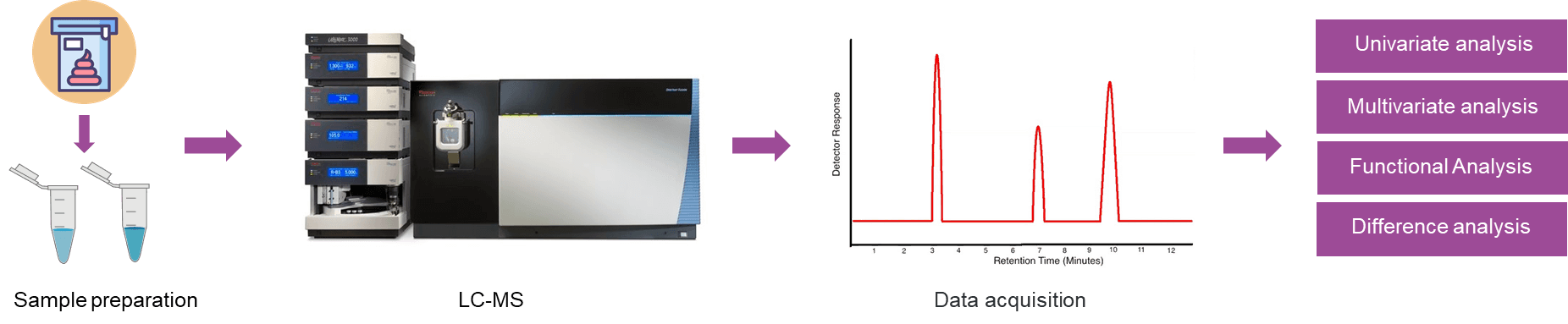 Fig 1. The workflow of untargeted metabolomics analysis
Fig 1. The workflow of untargeted metabolomics analysis
Targeted metabolomics analysis:
Our metabolomics platform provides a complete analysis of metabolites in feces, including short-chain fatty acids, amino acids and microbial metabolites.
- Mode: SRM / MRM
- Analysis content: Standard curve creation. Raw data preprocessing. Absolute quantification of metabolites.
- Detectable substances include but are not limited to:
Short chain fatty acids
Formate
Propionate
Valerate
Acetate
Butyrate
Isobutyrate
Isovalerate | Amino acids
Lysine
Alanine
Glycine
Glutamate
Methionine
Tyrosine
Phenylalanine
Valine
Leucine
Isoleucina | Microbial metabolism
TMAO
Dimethylamine | Glucose metabolism
Glucosa
Lactate
Succinate | Diet metabolism
Xylose
Galactose |
Pyrimidine metabolism
Uracil
Uridine | Purine metabolism
Oxypurinol
Hypoxanthine
Inosine
2-deoxyinosine | Others
Creatine
Creatinine
Malonato
Choline
3-phenylpropionate |
|
|
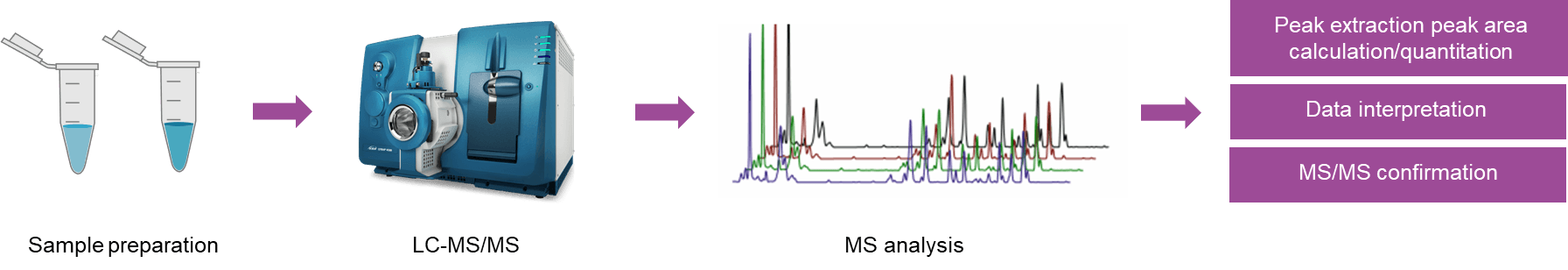 Fig 2. The workflow of target metabolomics analysis
Fig 2. The workflow of target metabolomics analysis
Sample Requirements
- Rat feces: ≥ 15-20 mg dry weight
- Mouse feces: ≥ 15-20 mg dry weight
- Human feces: a single histology of 200 mg or more is recommended.
Collection of stool samples in a sterile stool collector or other sterile utensils is required. Care should be taken to label the sample immediately and store it at low temperature, trying not to expose it to air for too long to avoid contamination and degradation. Before freezing the collected fresh stool, it is recommended to mix the sample with a sterile stick.
Deliverables
- Experimental procedure
- Parameters of liquid chromatography / gas chromatography and MS
- MS raw data files and MS data quality checks
- Bioinformatics analysis report
Turnaround time: 1-4 weeks
Bioinformatics Analysis
| Standard analysis | 1. Data quality control (QC correlation analysis, PCA analysis of total samples) |
| 2. Qualitative and quantitative results of metabolites |
| 3. Differential metabolite screening (PCA, PLS-DA dimensionality reduction treatment, volcano map) |
| 4. Differential metabolite analysis: Venn diagram, cluster heat map, metabolite correlation analysis, Z-score analysis, pathway analysis, ROC analysis |
| Personalized analysis | Weighted gene co-expression network analysis (WGCNA) |
| One way ANOVA |
Creative Proteomics provides metabolomics services for feces samples, and will deliver accurate and detailed data and analysis reports to you. If you want to detect other metabolites but have not found them, you can tell us through the inquiry form, and our technicians will communicate with you.
For Research Use Only. Not for use in diagnostic procedures.

 Fig 1. The workflow of untargeted metabolomics analysis
Fig 1. The workflow of untargeted metabolomics analysis Fig 2. The workflow of target metabolomics analysis
Fig 2. The workflow of target metabolomics analysis