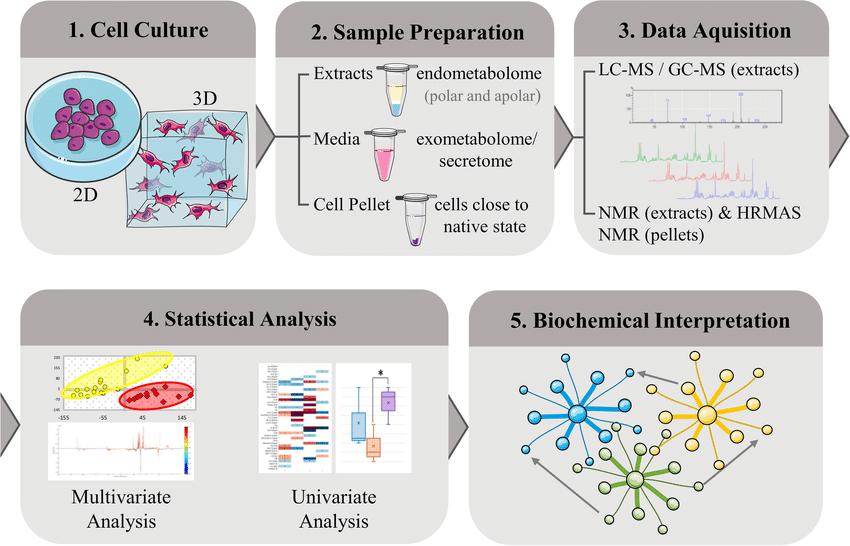
Metabolomics is the large-scale analysis of small-molecule metabolites that reflect the real-time physiological state of biological systems. It enables researchers to connect genotype to phenotype, revealing how cells respond to disease, treatment, or environmental changes.
But modern metabolomics isn't just about generating data—it's about biological clarity, mechanism-focused analysis, and fast, confident decision-making. Whether you're exploring metabolic phenotypes, validating pathway shifts, or integrating multi-omics, what you need is actionable, well-annotated, and publication-ready results—with no guesswork.
That's why our metabolomics services are designed from your research goals backward:
- Need to explore metabolism without assumptions? → We provide full-spectrum untargeted metabolomics
- Need robust quantification for validation or submission? → We deliver targeted metabolomics with full LOD/LOQ/QC
- Need to interpret results at the pathway level? → We offer curated, mechanism-based annotation, not generic lists
Why Choose Creative Proteomics for Metabolomics?
Metabolomics Solutions Tailored to Your Research Goals
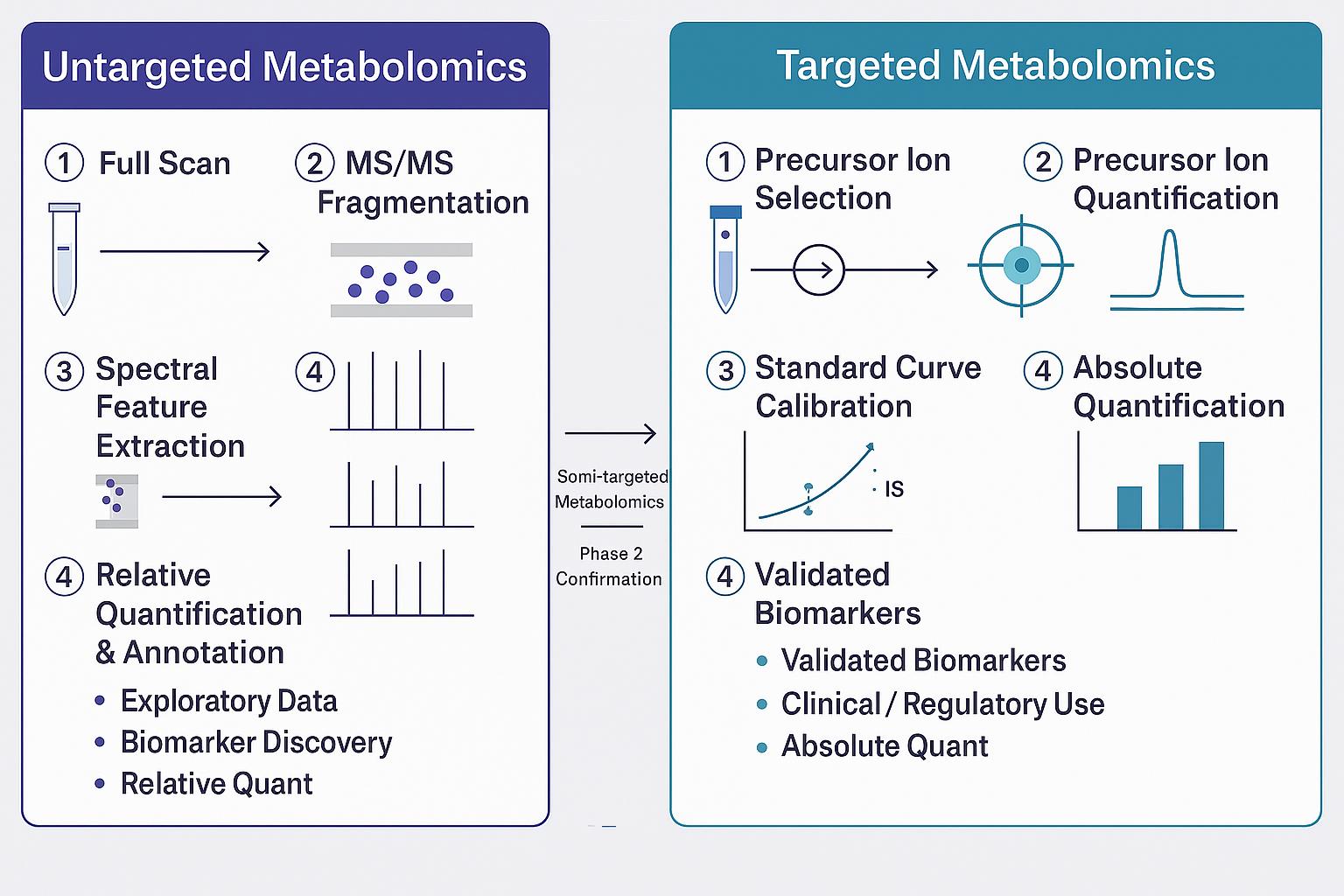
Untargeted metabolomics provides a global, hypothesis-generating view of small molecule metabolites in biological systems. This approach is ideal for:
- Discovering novel metabolites
- Identifying altered metabolic pathways
- Understanding phenotypic or condition-specific metabolic changes
We detect and relatively quantify hundreds to thousands of metabolites across sample types. Suitable for early-stage research, comparative studies, and systems biology applications.
Targeted metabolomics focuses on quantifying a predefined set of metabolites with high accuracy and sensitivity. It is hypothesis-driven, often used for:
- Validating biomarkers
- Monitoring treatment effects
- Studying specific pathways in-depth
Our targeted metabolomics panels cover a wide range of compound classes and are customizable for human, plant, and microbial studies. All measurements are based on internal/external standards with LOD/LOQ and QC validation.
Untargeted vs. Targeted Metabolomics: How to Choose the Right Approach?
| Aspect |
Untargeted Metabolomics |
Targeted Metabolomics |
| Purpose |
Explore unknown metabolites and generate hypotheses |
Precisely quantify known metabolites |
| Approach |
Broad, unbiased screening |
Focused, hypothesis-driven analysis |
| Analyte Scope |
Hundreds to thousands of features |
Dozens to hundreds of predefined compounds |
| Quantification |
Relative quantitation |
Absolute quantitation (internal standards, calibration curves) |
| Sensitivity |
Moderate |
High |
| Typical Use Cases |
Early discovery, pathway screening, phenotype comparison |
Biomarker validation, drug response, mechanistic studies |
| Flexibility |
High – suitable for unknowns |
Fixed or customizable panels |
| Data Output |
Feature table, MS/MS annotations, PCA, volcano plot, pathway enrichment |
Concentration tables, LOD/LOQ, CV%, validation metrics |
| Recommended When... |
You don't know what to expect and want full coverage |
You have clear targets and need robust quantitation |
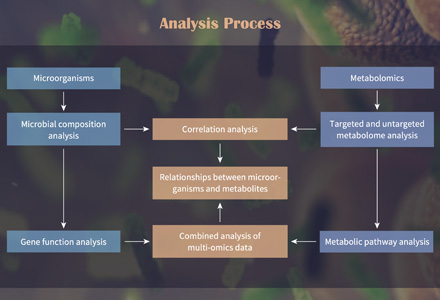
Our bioinformatics team supports pathway-level interpretation through advanced statistical modeling and visual analysis. Services include:
- Metabolite-to-pathway annotation (KEGG, Reactome, HMDB)
- Differential abundance analysis (volcano plots, heatmaps, PCA/PLS-DA)
- Pathway enrichment and network topology insights
- Biological interpretation & regulatory mechanism identification
Understand not only what changes, but how fast metabolites flow through biological pathways. Creative Proteomics offers 13C-labeled metabolic flux analysis, enabling:
- Dynamic quantification of pathway activity
- Mechanistic insights into cellular metabolism
- Integration with metabolomics and transcriptomics for network-level modeling
Available as an add-on for microbial, plant, or mammalian cell systems. Contact us to discuss tracer substrates, modeling approaches, and data integration options.
Metabolite and Pathway Coverage in Our Metabolomics Services
Metabolite Classes
| Category |
Subclasses / Examples |
| Amino Acids & Derivatives |
Amino acids (20), Biogenic amines, Polyamines, BCKA panel |
| Lipids & Related Metabolites |
Fatty acids (21), Free fatty acids, Carnitine & Acylcarnitine, β-oxidation flux |
| Organic Acids |
Organic acids (11), Ethanol metabolites |
| Carbohydrates & Derivatives |
Carbohydrate metabolism products (26), Glycan metabolites, Glycogenesis metabolites, Gluconeogenesis intermediates |
| Vitamins & Cofactors |
Cofactors & vitamins (12), Nicotinamide adenine dinucleotide (NAD/NADH), Redox cofactors |
| Plant Secondary Metabolites |
Flavonoids (17), Isoflavonoids, Dihydroflavanones, Puerarin, Anthocyanins (6), Carotenoids (10), Terpenoids (11), Polyphenols, Stilbenes (3), Tannins (2), Iridoids (5), Lignans (4), Coumarins (3), Alkaloids (8), Polyketides, Quinones, Saponins (2), Glucosinolates (5), Indoles & indole-sulfur compounds (5), Plant hormones (11), Plant lectins (4) |
| Pigments & Functional Molecules |
Chlorophyll (6), Glutathione, Oxidative stress markers |
| Toxins & Exogenous Compounds |
Toxins(4), Xenobiotic metabolites, Glyphosate & metabolites |
| Specialized Metabolites |
Urolithin A, Trimethylamine oxide (TMAO), Reactive oxygen species (ROS), Animal hormones |
Metabolic Pathways
| Category |
Examples |
| Central Carbon Metabolism |
Central carbon metabolism, Glycogenesis, Gluconeogenesis |
| Amino Acid Metabolism |
Urea cycle metabolism, Methionine cycle metabolism, One-carbon metabolism, BCKA metabolism |
| Energy Metabolism |
Energy metabolism, NAD⁺/NADH balance, β-oxidation flux |
| Lipid Metabolism |
Lipid metabolism, Acylcarnitine metabolism, Oxylipin metabolism |
| Carbohydrate Metabolism |
Carbohydrate metabolism, Glycan metabolism |
| Nucleotide Metabolism |
Nucleotide metabolism (2) |
| Plant-Specific Pathways |
Plant hormone pathways, Secondary metabolite pathways |
| Redox & Stress Response |
ROS metabolism, Redox cofactors, Glutathione metabolism, Oxidative stress markers |
| Environmental & Toxicology |
Xenobiotic metabolism, Glyphosate pathway |
| Other Specialized Pathways |
Bile acid metabolism, Ethanol metabolism, Urolithin A metabolism, TMAO metabolism |
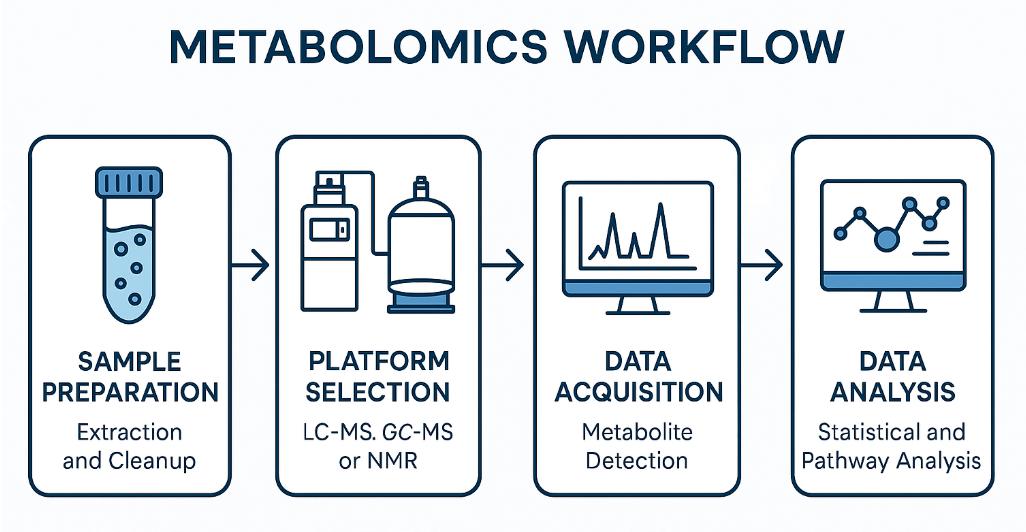
What Instruments Do We Use for Metabolomics?
At Creative Proteomics, we utilize a full suite of LC-MS and GC-MS platforms, enabling high-resolution, high-throughput metabolomics across diverse sample types. Our systems support both untargeted discovery and targeted quantitation workflows with precision, reproducibility, and wide chemical class coverage.
Our platforms are optimized for:
- High-resolution metabolite profiling for global untargeted analysis
- Triple quadrupole–based quantitation for accurate, targeted panels
- Flexible separation (UPLC and GC) tailored to compound polarity and volatility
- Dual-mode ionization (positive/negative) for enhanced metabolite class detection
- Automated QC workflows following strict SOPs and method validation guidelines
Key Technical Parameters
| Metric |
Typical Capability |
| Mass Accuracy |
≤ 5 ppm (HRMS) |
| LOD/LOQ |
< 1 nM (compound-dependent) |
| Dynamic Range |
104–106 |
| Reproducibility (QC CV%) |
< 15% |
| Ionization Modes |
Positive & Negative (ESI/APCI) |
| Quantitation Types |
Absolute (w/ standards) or Relative |
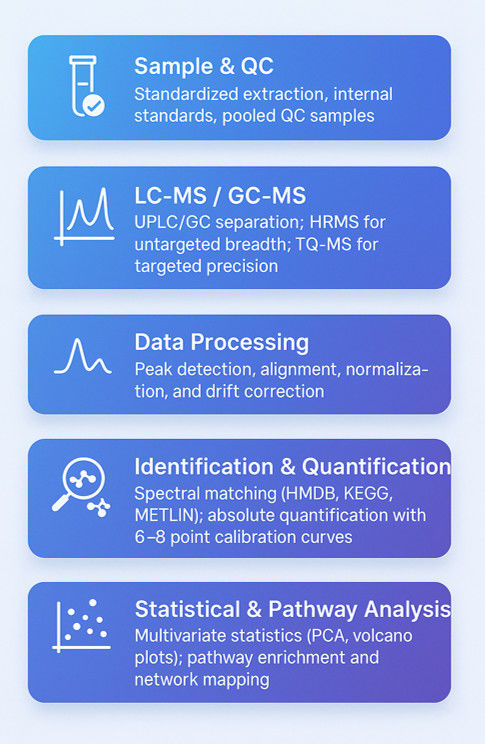
Metabolomics Analysis Workflow — A Step-by-Step Guide
How to Submit Samples for Metabolomics Analysis
Please refer to the table below for accepted sample types, minimum volume/weight, and handling instructions. If you are working with a custom matrix or rare sample type, contact our support team for consultation.
| Sample Type |
Recommended Volume / Weight |
Storage Conditions |
Shipping Notes |
| Plasma / Serum |
≥ 100–200 µL |
−80°C |
Ship on dry ice, avoid freeze–thaw |
| Urine |
≥ 500 µL–1 mL |
−80°C |
Filter or centrifuge if needed |
| Tissues (Animal/Plant) |
≥ 30–50 mg |
Snap-frozen, −80°C |
Pulverize if possible; ship in cryovials |
| Cultured Cells |
≥ 1–5 million cells |
Pellet form, −80°C |
Wash with PBS, remove media |
| Feces / Gut Microbiota |
≥ 100 mg |
−80°C |
Store in sterile tubes or fecal collection kits |
| CSF / Bile / Saliva |
≥ 100 µL |
−80°C |
Special precautions may apply (low protein samples) |
| Culture Supernatant / Media |
≥ 1 mL |
−80°C |
Pre-clear if possible, avoid additives |
| Soil / Environmental |
≥ 100 mg |
−20°C or −80°C |
Dry/freeze-dry recommended |
| Plant Extracts |
≥ 500 µL |
−80°C |
Record extraction solvent and method |
General Handling Notes
- Avoid adding preservatives, antibiotics, or buffer salts unless specified
- Store samples in leak-proof cryovials, preferably 2 mL or smaller
- For liquid samples: centrifuge and aliquot to avoid repeated thawing
- All samples must be shipped with sufficient dry ice for multi-day transit
Deliverables: What You Receive from Metabolomics Analysis
Data Files
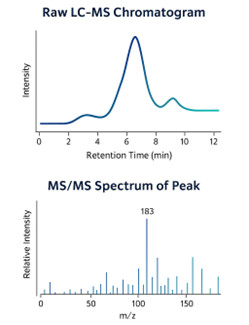
- Raw mass spectrometry data (vendor format + open formats: mzML/mzXML)
- Processed feature tables with m/z, retention time, intensity
Quantification & Identification
- Relative (untargeted) or absolute (targeted)quantification results
- Metaboliteidentification with IDs, MS/MS match scores, and confidence levels
Quality Control Report
- Internal/external standard performance
- QC reproducibility, CV values, and drift correction plots
Statistical & Pathway Analysis
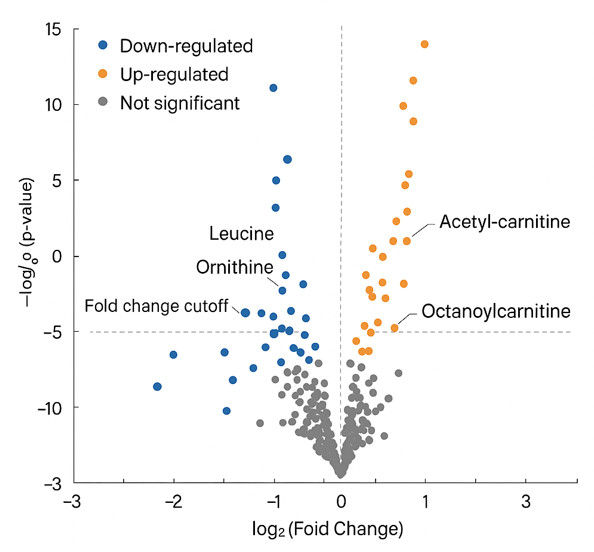
- PCA, PLS-DA, volcano plots, and heatmaps
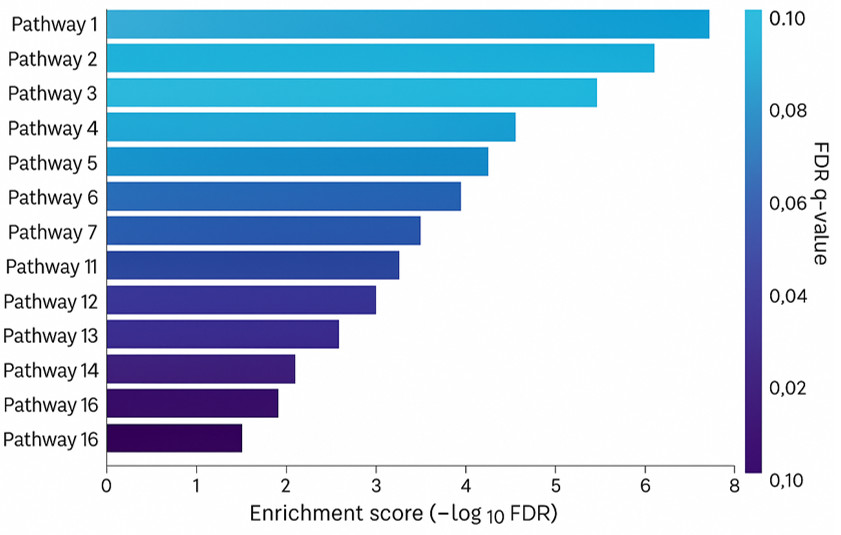
- Pathway enrichment maps and differential metabolite lists
Final Report
- Methods and QC summary
- Highlighted key metabolites and pathways
Hot Analytical Metabolite Categories
Applications of Metabolomics Service
Creative Proteomics's metabolomics service offers advanced solutions for exploring and analyzing metabolites, providing valuable insights into diverse areas of research.
Comprehensive profiling and focused study of metabolites in biofluids such blood, urine, and cerebrospinal fluid are made possible by metabolomics. Researchers can find potential biomarkers linked to particular diseases or physiological situations by examining the metabolite patterns.
Metabolomics, for instance, can assist in finding distinctive chemical signatures that distinguish between healthy people and cancer patients in cancer research. These biomarkers can help with early detection, prognosis evaluation, and therapy response tracking.
Metabolomics offers insights into drug metabolism, efficacy, and toxicity. Creative Proteomics's metabolomics service enables researchers to study the metabolic effects of drugs and identify potential drug targets. By analyzing the metabolomic profiles of cells, tissues, or biofluids after drug exposure, researchers can gain a deeper understanding of drug mechanisms of action and evaluate drug safety and efficacy.
Metabolomics can assist in identifying drug metabolites, elucidating metabolic pathways involved in drug metabolism, and predicting drug-drug interactions.
By making it easier to analyze the metabolite profiles of food samples, metabolomics enables researchers to determine the nutritional value, assess the quality of the food, and investigate how different food preparation methods affect metabolite profiles.
Understanding nutrition consumption and metabolism, finding bioactive substances in food, and assessing the impacts of additions or pollutants can all be done with the aid of metabolomics. Understanding the link between diet and health, creating individualized nutrition plans, and assuring the safety and quality of food products are all made easier with the use of this knowledge.
Metabolomics enables the analysis of metabolomic profiles in organisms exposed to environmental stressors or pollutants. By studying the metabolic responses of organisms to environmental changes, researchers can assess the impact on their metabolism and identify potential biomarkers of exposure or effect.
Metabolomics can help in understanding the biochemical pathways involved in the detoxification and metabolism of environmental contaminants. It aids in predicting ecological risks, assessing the effects of pollutants on human health, and contributing to environmental protection efforts.
Plant metabolite profiles may be analyzed more easily due to metabolomics, which reveals details about how plants use their nutrients and respond to biotic and abiotic challenges.
Understanding the biochemical pathways involved in crop development, finding metabolites linked to desired features, and optimizing agricultural techniques for better yield and quality are all possible with the help of metabolomics. This knowledge aids in the development of sustainable agricultural methods, the improvement of crop resistance to diseases, and the breeding of improved crop types.
Systems biology approaches are supported by metabolomics by providing the essential analytical tools and knowledge. Researchers can obtain a comprehensive understanding of biological processes and their regulation by integrating metabolomics data with other "omics" technologies including genomes, transcriptomics, and proteomics. Discovering new metabolic pathways, locating important regulatory nodes, and figuring out how biological phenomena are caused are all made easier by metabolomics. This holistic strategy can be used to model diseases, comprehend cellular metabolism, and find possible treatment targets.
Reference
- Shi, Zhan, et al. " A comprehensive mass spectrometry-based workflow for clinical metabolomics cohort studies." Metabolites 12.12 (2022): 1168.
Case: Metabolic Dysfunction in Young Adulthood Predicts Long-Term Cardiovascular Outcomes: Insights from a Multi-Cohort Study
Background
Understanding the antecedents of cardiovascular risk in young adults is crucial for effective prevention. Traditional risk assessment methods may not capture early metabolic changes associated with cardiovascular diseases (CVD). This study aims to identify specific metabolic features in young adulthood that predict an adverse cardiovascular phenome over two decades, utilizing advanced analytical techniques.
Sample
The study includes 2330 individuals from the CARDIA cohort (mean age 32.1±3.6 years; 45% women; 45% Black). Additionally, validation is performed on 1898 participants from the FHS Offspring Cohort (mean age at metabolite measurement 54.9±9.7 years).
Methods
Statistical Methods:
- Graphical examination of distributions of continuous subclinical endpoints and covariates.
- Transformation of variables, including logarithmic and hyperbolic arcsine transformations.
- Pooled cohort equation risk calculation and standardization of continuous endpoints and covariates.
- Metabolite imputation and log transformation, followed by correlation analysis and PCA.
Single Metabolite Regressions:
- Linear or logistic regression for each subclinical CVD outcome against each metabolite.
- Three sets of models: unadjusted, adjusted for demographic factors, and adjusted for clinical risk factors.
- Application of Benjamini-Hochberg false discovery rate control.
Elastic Net and PCA:
- Two-phase regression approach using elastic net regression and subsequent PCA to identify metabolites linked to subclinical CVD endpoints.
- Generation of vascular and myocardial scores based on metabolite weighting and concentration.
- Survival analysis using Cox regression with adjustment for clinical factors.
FHS Validation:
- Validation of identified metabolites and scores in the FHS cohort using linear regressions.
- Examination of the association with incident CVD and death.
Pathway Analysis:
- Detailed pathway analysis to explore the underlying mechanisms of metabolic dysfunction.
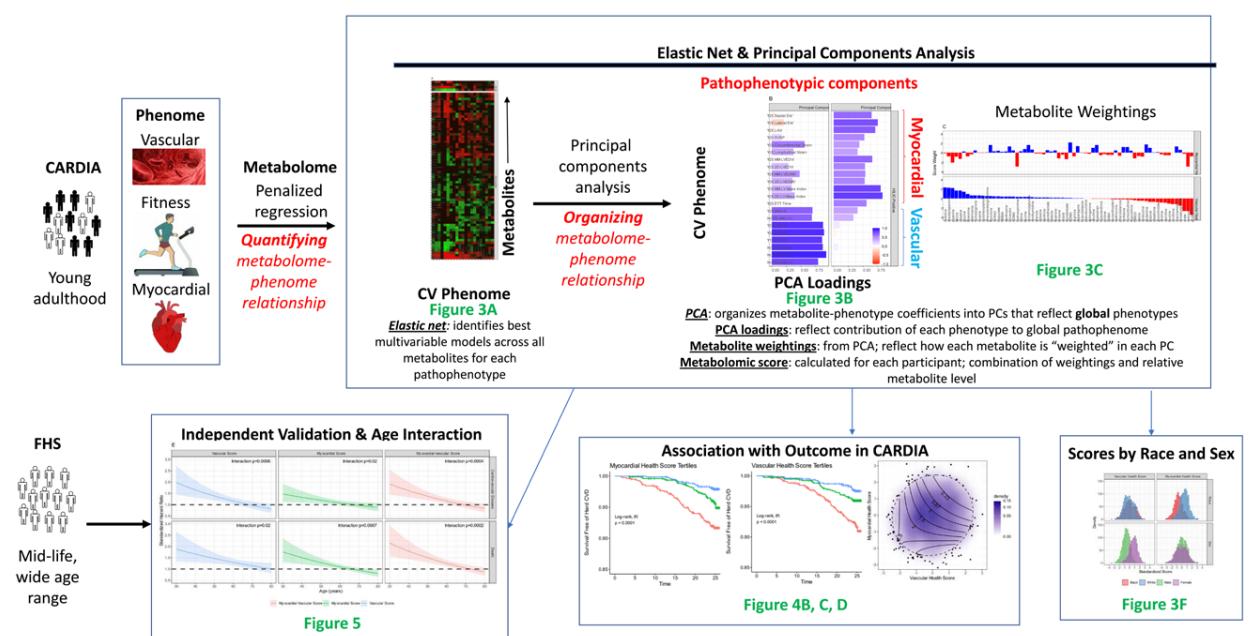 Study scheme.
Study scheme.
Results
Clinical Characteristics:
- Description of demographic and clinical characteristics of the study populations.
- Comparison between the CARDIA and FHS cohorts.
Metabolic Dysfunction Associations:
- Associations between metabolites in young adulthood and subsequent development of adverse cardiovascular phenome.
- Network in silico approach revealing pathways implicated in myocardial and vascular remodeling.
Metabolic Scores and Cardiovascular Risk:
- Generation of vascular and myocardial health scores associated with specific metabolites.
- Scores demonstrated associations with future CVD, and the association was stronger in young adulthood.
Validation and Age-Dependent Association:
- Replication of findings in the FHS cohort.
- Strong interaction between age and metabolic scores in predicting CVD, with the highest hazard in young adulthood.
Race Differences and Clinical Relevance:
- Identification of race differences in myocardial health scores.
- Emphasis on the clinical relevance of precise metabolic measures in early adulthood for predicting long-term cardiovascular outcomes.
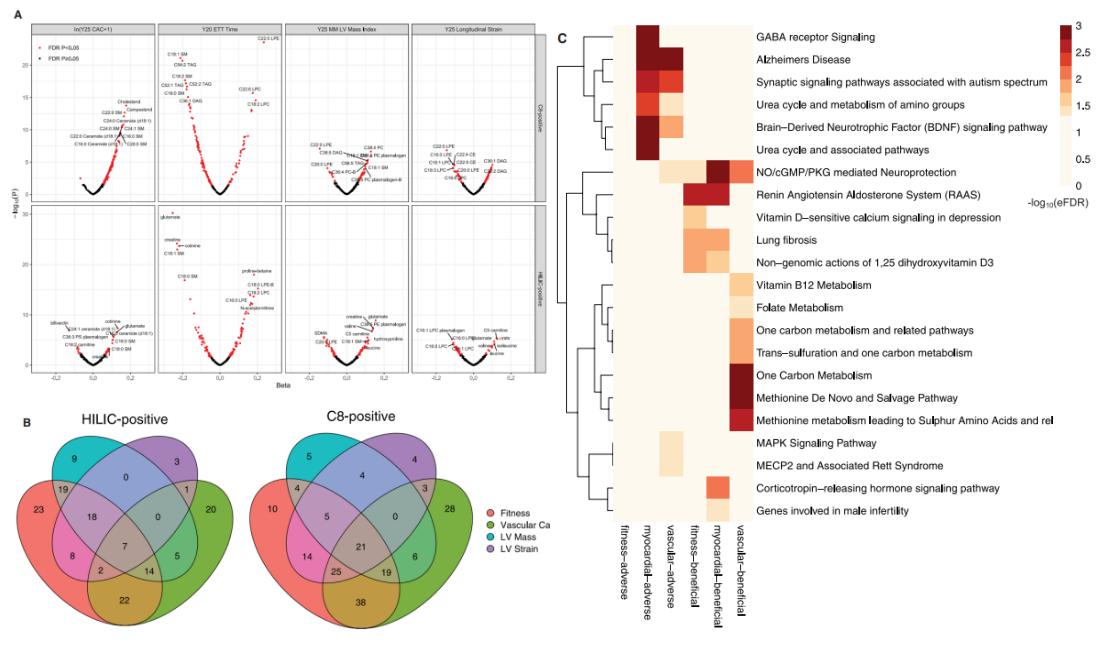 Metabolites in early adulthood are associated with the cardiovascular phenome and identify pathways central to cardiovascular disease development.
Metabolites in early adulthood are associated with the cardiovascular phenome and identify pathways central to cardiovascular disease development.
Reference
- Murthy, Venkatesh L., et al. " Comprehensive metabolic phenotyping refines cardiovascular risk in young adults." Circulation 142.22 (2020): 2110-2127.
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566











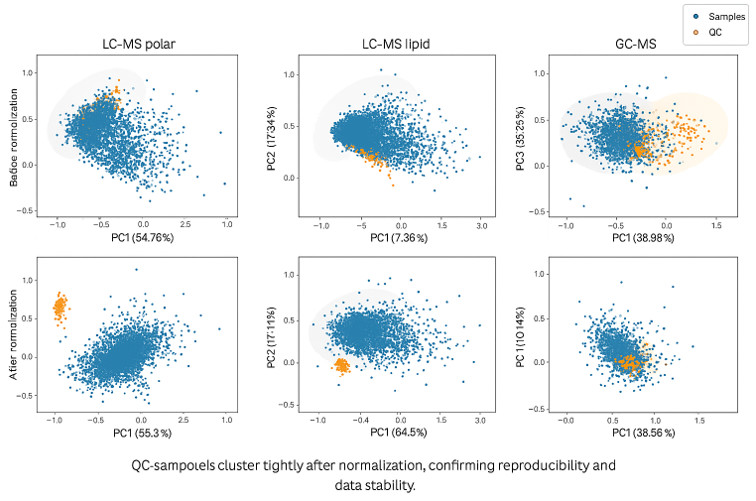





 Study scheme.
Study scheme. Metabolites in early adulthood are associated with the cardiovascular phenome and identify pathways central to cardiovascular disease development.
Metabolites in early adulthood are associated with the cardiovascular phenome and identify pathways central to cardiovascular disease development.