Oxaloacetic Acid Analysis Service
Submit Your InquiryWhat is Oxaloacetic Acid?
Oxaloacetic acid (also known as oxalacetic acid or OAA) is a four-carbon dicarboxylic acid that plays an essential role in several metabolic pathways, including gluconeogenesis, amino acid metabolism, and the citric acid cycle. OAA is synthesized from pyruvate by the enzyme pyruvate carboxylase and is subsequently converted to phosphoenolpyruvate by the enzyme phosphoenolpyruvate carboxykinase, which is an essential step in gluconeogenesis. OAA can also be converted to aspartate, which is a precursor for several amino acids, including arginine, methionine, and lysine. OAA is also involved in the malate-aspartate shuttle, which transfers reducing equivalents across the mitochondrial membrane in oxidative phosphorylation.
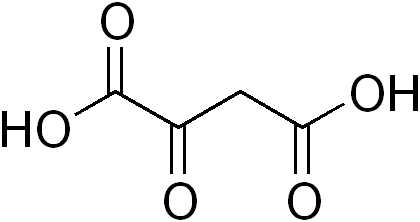 Molecular structure of oxaloacetic acid
Molecular structure of oxaloacetic acid
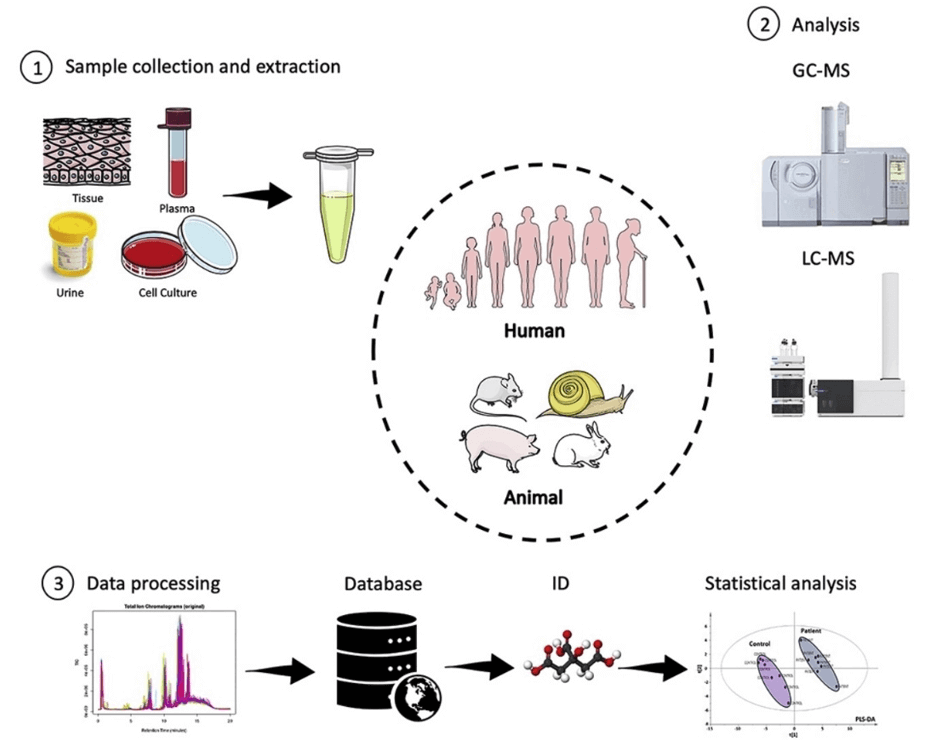
Creative Proteomics offers a variety of services to support the analysis of oxaloacetic acid metabolism, including sample preparation, LC-MS analysis, and data analysis.
Oxaloacetic acid Analysis Technology Platform in Creative Proteomics
LC-MS is the most commonly used chromatography-based method for the analysis of oxaloacetic acid. In LC-MS, a liquid chromatography system is used to separate OAA from other compounds in the sample, which is then detected by a mass spectrometer.
The stationary phase material of the liquid chromatography system is selected as a column filled with C18 or C8. After separation by liquid chromatography, the compounds are introduced into a mass spectrometer, where they are ionized and detected. The mass spectrometer measures the mass-to-charge ratio (m/z) of the ions, providing information about the identity and abundance of the compounds in the sample.
LC-MS can perform OAA analysis in several modes, including selected ion monitoring (SIM) and multiple reaction monitoring (MRM). SIM mode measures specific m/z values of OAA ions, while MRM mode measures two specific m/z values corresponding to OAA precursor and product ions. MRM mode has higher sensitivity and specificity than SIM mode and is the preferred OAA analysis mode.
To ensure accurate and precise quantification, stable isotope-labeled internal standards are commonly used in the analysis of OAA by LC-MS. The internal standard has a chemical structure similar to that of OAA but contains a stable isotope label that allows accurate quantification and correction for variations in sample preparation and instrument performance.
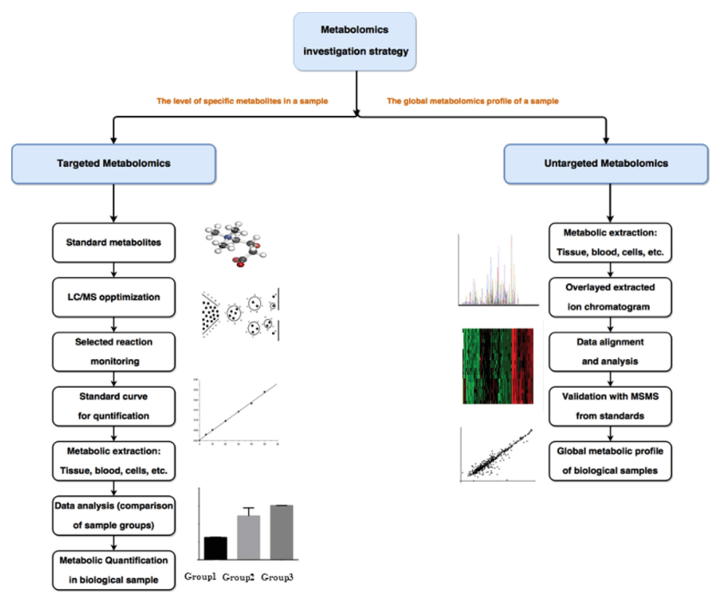
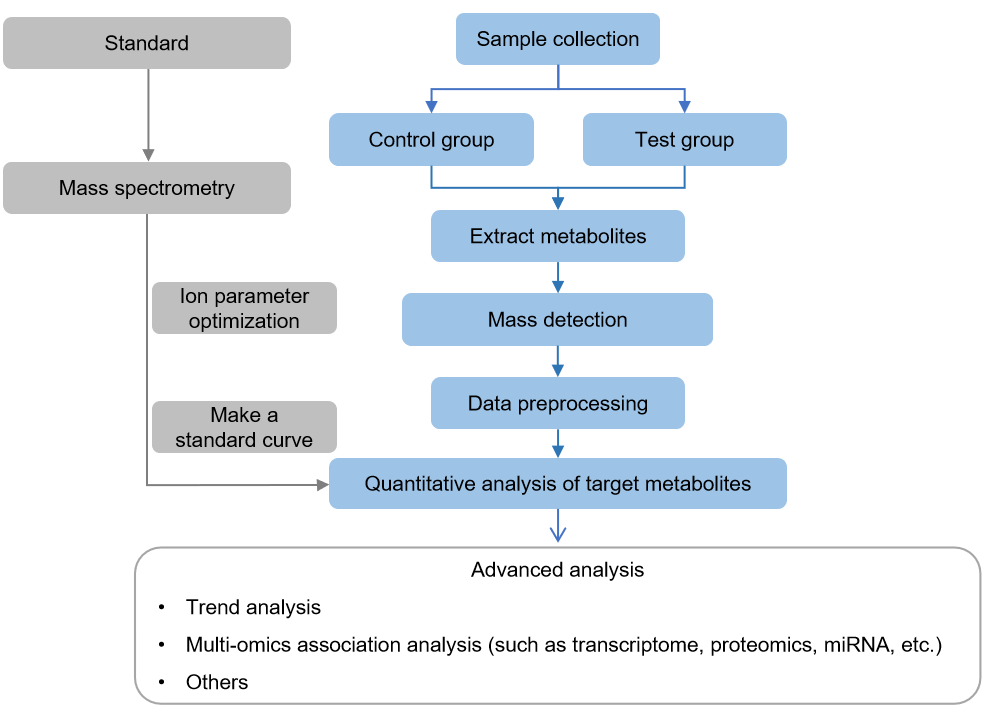
Technical Route of Targeted Metabolomics of Glucose
Feedback to Customers
Creative Proteomics will provide you with detailed technical reports, including
- Experimental steps
- Related mass spectrometry parameters
- Part of the mass spectrum picture
- Raw data
- Metabolic molecular identification results
Creative Proteomics offers several approaches to metabolomics studies, delivers precise and detailed data and analysis report. We can also customize the methods or establish new methods together with our collaborators, so they are fit-for-purpose and meet your specific needs. If you have any questions or specific requirements, please feel free to contact us.
References
- Riera-Borrull M, Rodríguez-Gallego E, Hernández-Aguilera A, et al. Exploring the process of energy generation in pathophysiology by targeted metabolomics: performance of a simple and quantitative method. Journal of the American Society for Mass Spectrometry, 2015, 27(1): 168-177.
- Kim M J, Lee M Y, Shon J C, et al. Untargeted and targeted metabolomics analyses of blackberries – Understanding postharvest red drupelet disorder. Food Chemistry, 2019, 300:125169.
- Wang X, Zhao X, Zhao J, et al. Serum metabolite signatures of epithelial ovarian cancer based on targeted metabolomics. Clinica Chimica Acta, 2021, 518: 59-69.