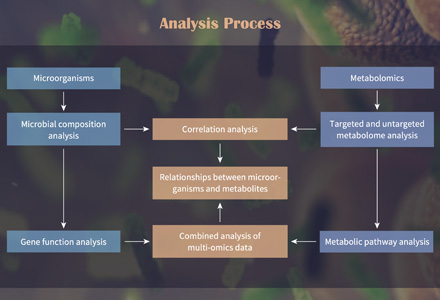
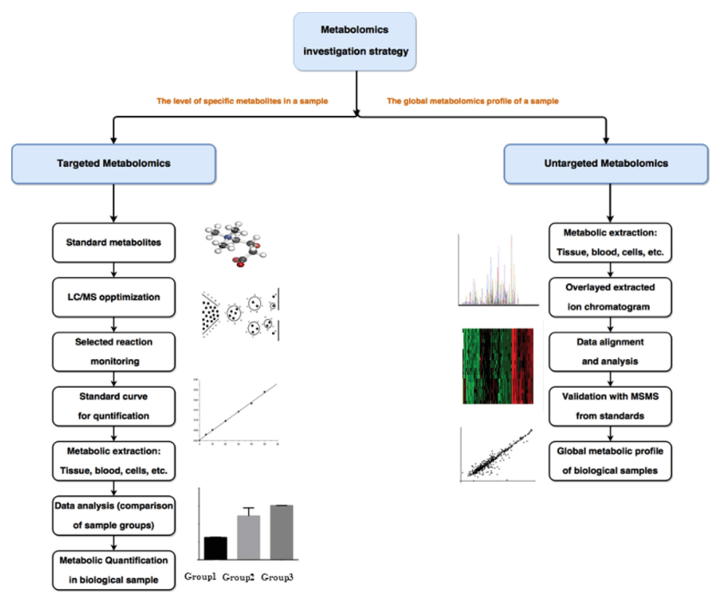
What Are Organic Acids and Why Analyze Them?
Organic acids are small, acidic metabolites with one or more carboxyl groups, including citric, malic, lactic, fumaric, succinic, and acetic acids. They are central nodes in energy and carbon metabolism rather than peripheral by-products.
In metabolism, organic acids mainly act as:
- TCA-cycle intermediates (citric, malic, fumaric, succinic, oxaloacetic acids) that reflect mitochondrial flux and ATP production.
- Glycolytic and fermentation products (lactate, pyruvate, acetic, butyric acids) that report on oxygen status, mitochondrial function, and microbial fermentation.
- Signaling metabolites such as itaconic acid that influence immune activation and inflammatory pathways.
In food and agriculture, organic acids are key markers of taste, acidity, freshness, fermentation progress, and product authenticity.
Because they sit close to core metabolic flux, small changes in organic acid profiles can indicate major pathway shifts. High-quality organic acid data help you:
- Link gene or protein changes to real metabolic outcomes.
- Separate on-target metabolic effects from off-target toxicity.
- Track how diet, microbiota, environment, or treatments reshape metabolic networks.
Targeted LC–MS/MS organic acids analysis provides the sensitivity and selectivity needed to capture these changes across complex matrices such as plasma, urine, tissues, feces, cell extracts, foods, and environmental samples.
What Questions Can Organic Acids Analysis Answer?
Organic acids profiling is most informative when it is linked to clear biological or product questions. Our organic acids analysis service commonly supports projects such as:
Targeted Organic Acids Panel and Pathway Coverage
Our targeted organic acids panel is organized by pathway and application so you can quickly check whether your metabolites of interest are covered.
| Panel category |
Organic acids in this panel (include but are not limited to)* |
| TCA & central carbon organic acids |
Citric acid, Isocitric acid, cis-Aconitic acid, trans-Aconitic acid, α-Ketoglutaric acid, Succinic acid, Fumaric acid, Malic acid, Oxaloacetic acid, Pyruvic acid, Lactic acid, Phosphoenolpyruvic acid (PEP), 2-Oxobutyric acid, 2-Oxoisovaleric acid, Gluconic acid, Glyceric acid |
| SCFAs & fermentation organic acids |
Acetic acid, Propionic acid, Butyric acid, Isobutyric acid, 2-Methylbutyric acid, Valeric acid, Isovaleric acid, Caproic (Hexanoic) acid, Heptanoic acid, Lactic acid (D/L), Formic acid, Succinic acid, Pyruvic acid, 3-Hydroxybutyric acid, Acetoacetic acid, Fumaric acid |
| Metabolic disease–related organic acids |
3-Hydroxybutyric acid, Acetoacetic acid, Adipic acid, Suberic acid, Sebacic acid, Glutaric acid, Methylmalonic acid, Succinylacetic acid, Hippuric acid, Uric acid, Oxalic acid, Glyoxylic acid, Lactic acid, Pyruvic acid, Citric acid |
| Immunometabolism & signaling organic acids |
Itaconic acid, cis-Aconitic acid, Fumaric acid, 2-Hydroxyglutaric acid (D/L or total), Lactic acid, Pyruvic acid, Succinic acid, Kynurenic acid, Quinolinic acid |
| Plant & food-related organic acids |
Malic acid, Citric acid, Tartaric acid, Quinic acid, Shikimic acid, Ascorbic acid (Vitamin C), Gallic acid, Caffeic acid, Chlorogenic acid, Benzoic acid, Syringic acid, Ferulic acid, Succinic acid, Acetic acid, Oxalic acid |
| Soil & environmental organic acids |
Oxalic acid, Malonic acid, Maleic acid, Succinic acid, Fumaric acid, Citric acid, Tartaric acid, Gluconic acid, Formic acid, Acetic acid, Propionic acid, Benzoic acid, Humic-related acids, Fulvic-related acids |
*The panel is periodically optimized. Additional organic acids can be included or added as custom targets upon request.
Why Choose Creative Proteomics for Organic Acids Analysis?
- Pathway-centered organic acids panel: Coverage across TCA cycle, central carbon metabolism, SCFAs, plant/food acids, and signaling metabolites.
- Advanced LC–MS/MS platform: High-sensitivity QTRAP®-based workflows with optimized MRM methods for organic acids.
- Matrix experience across biology, food, and environment: Proven protocols for plasma, urine, tissues, feces, foods, beverages, and environmental samples.
- Robust QC and method validation: Calibration, QC samples, and internal standards to ensure reproducible quantification.
- Integration with broader metabolomics strategies: Organic acids panels that can be combined with other targeted or untargeted metabolomics services.
- Project-driven data interpretation: Reports and visualizations tailored to your experimental design and decision points, not just generic output.
Analytical Platform for LC–MS/MS Organic Acids Quantification
Core Instrumentation
LC–MS/MS system – High-performance triple quadrupole platform (e.g., AB Sciex QTRAP® 6500 or equivalent) with electrospray ionization and optimized MRM methods for organic acids.
UHPLC system – Binary or quaternary UHPLC with precise autosampler injection for stable gradients and low carryover in polar metabolite analysis.
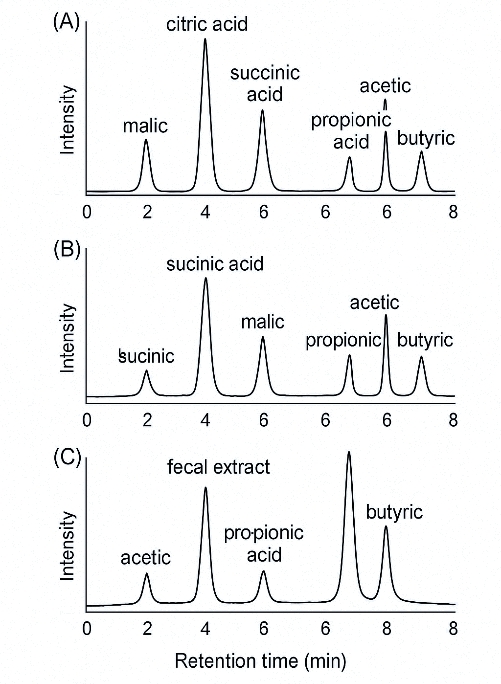
LC columns – Reversed-phase C18 and/or HILIC columns selected to retain and separate highly polar and isomeric organic acids.
Sample preparation tools – Refrigerated centrifuges, vortex mixers, and homogenizers enabling cold protein precipitation and extraction under controlled conditions.
Data processing & QC – Vendor LC–MS/MS software combined with in-house pipelines for MRM integration, calibration, QC tracking, and downstream statistical and pathway analysis.
Key Analytical Parameters
| Parameter |
Typical configuration / range* |
Why it matters for your project |
| Detection mode |
LC–MS/MS, ESI, Multiple Reaction Monitoring (MRM) |
Targeted, high-sensitivity detection of organic acids |
| Polarity |
Mainly negative ESI; positive ESI as needed |
Optimized ionization for carboxylic acids and related metabolites |
| LC separation |
C18 and/or HILIC, ~0.2–0.4 mL/min |
Robust separation of polar, closely eluting organic acids |
| Run time per sample |
Typically ~10–20 min (method-dependent) |
Balances depth of coverage with sample throughput |
| Calibration & IS |
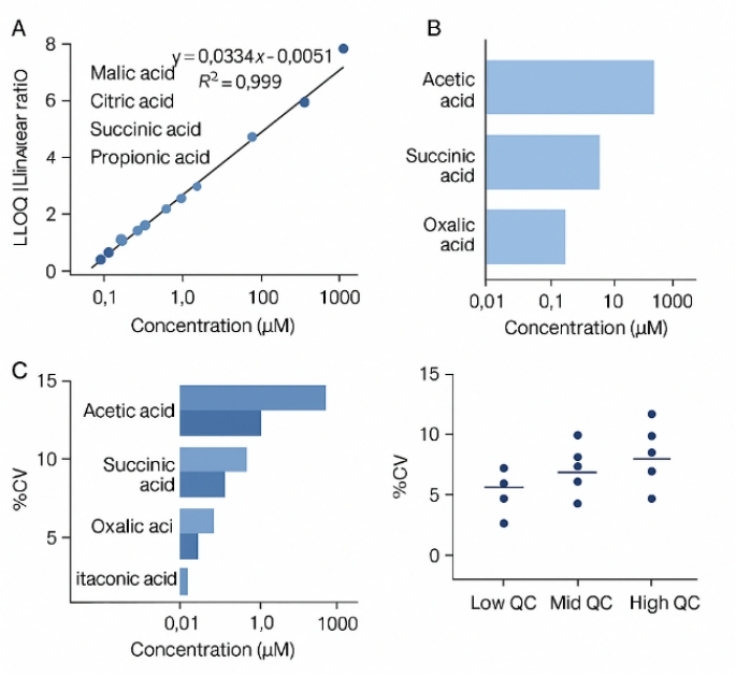
≥ 6-point calibration curves + isotope/structural internal standards |
Supports reliable quantitation across the validated range |
| QC design |
Blanks, system suitability, pooled QC samples per batch |
Monitors stability, carryover, and reproducibility |
| Typical performance |
Good linearity; QC %CV typically ≤ 15% |
Ensures data quality for downstream statistics and interpretation |
LC–MS/MS Workflow for Organic Acids Quantification
How to Prepare and Submit Your Samples
| Category |
Requirement |
| Accepted sample types |
Plasma/serum, urine, cell pellets, tissues, feces, culture media, food & plant matrices |
| Minimum amount / volume |
Plasma/serum: ≥ 50–100 µL
Urine: ≥ 100 µL
Cell pellets: ≥ 1–5 × 10⁶ cells
Tissues: ≥ 10–20 mg
Feces: ≥ 20–30 mg
Media/extracts: ≥ 200 µL |
| Storage |
Freeze at –80°C immediately after collection; avoid freeze–thaw cycles |
| Shipping |
Ship on dry ice; samples must remain fully frozen |
| Containers |
1.5–2 mL screw-cap tubes; leak-proof; secure sealing |
| Labeling |
Sample ID, matrix, species, collection date |
| Avoid |
Heparin plasma; preservatives/buffers; thawed samples; contaminated or insufficient volume |
Notes: For unusual matrices (fermented foods, environmental samples, plant exudates), send extra aliquots for method adaptation.
Project Deliverables for Organic Acids Analysis
- Sample receipt sheet (sample IDs, matrix, received volume and condition).
- LC–MS/MS method summary (organic acids panel used, matrix type, key settings and QC design).
- Result table for all quantified organic acids (per-sample concentrations with units, Excel/CSV).
- Calibration and QC summary (calibration range and R², QC sample results).
- Standard result report (key tables and basic comparison figures, in PDF/Word).
- Raw LC–MS/MS data files (provided if agreed in advance).
Prospective randomized, double-blind, placebo-controlled study of a standardized oral pomegranate extract on the gut microbiome and short-chain fatty acids
Sivamani, R. K., Chakkalakal, M., Pan, A., Nadora, D., Min, M., Dumont, A., ... & Chambers, C. J.
Journal: Foods
Year: 2023
Effect of arabinogalactan on the gut microbiome: A randomized, double-blind, placebo-controlled, crossover trial in healthy adults
Chen, O., Sudakaran, S., Blonquist, T., Mah, E., Durkee, S., & Bellamine, A.
Journal: Nutrition
Year: 2021
B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids
Sanchez, H. N., Moroney, J. B., Gan, H., Shen, T., Im, J. L., Li, T., ... & Casali, P.
Journal: Nature Communications
Year: 2020