Tyrosine Analysis Service
Submit Your InquiryOverview
Tyrosine (L-tyrosine, Tyr) is an important nutrient essential amino acid, which plays an important role in the metabolism, growth and development of humans and animals, and is widely used in medicine, food, feed and chemical industries. Tyrosine can be used as a raw material for amino acid infusion and amino acid compound preparations, and as a nutritional supplement. Tyrosine can be used as a raw material for the preparation of pharmaceutical and chemical products such as peptide hormones, antibiotics, melanin, p-hydroxycinnamic acid, p-hydroxystyrene, etc.
Creative Proteomics offers a simple, accurate and reproducible method for the determination of tyrosine and its derivatives by high performance liquid chromatography-UV/FLD gradient (HPLC-UV/FLD) and liquid chromatography-mass spectrometry (LC-MS). This method enables the determination of low levels of tyrosine and promotes the research and applications related to tyrosine.
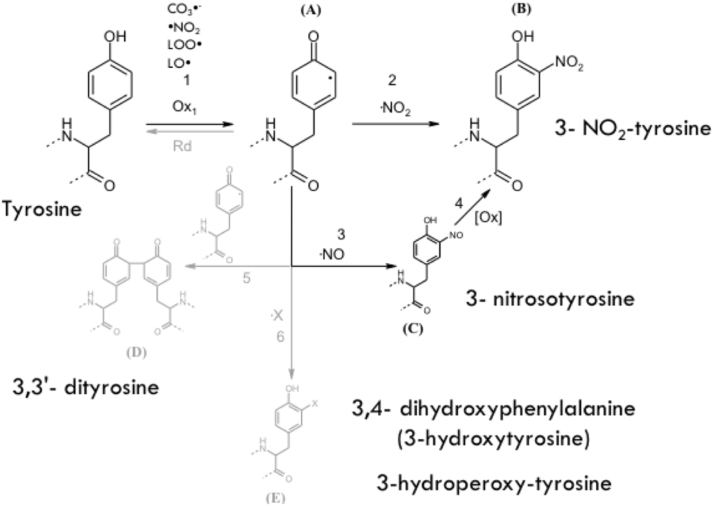 Figure 1. Tyrosine oxidation pathways. (Bartesaghi 2018).
Figure 1. Tyrosine oxidation pathways. (Bartesaghi 2018).
Applications of Tyrosine Analysis
- Determination and analysis of tyrosine and its derivatives
- Melanin improvement
- Quality control of food nutrition supplement
- The quality and safety of amino acid nutrition medicine in biochemical research
- Drug nutrient improvement
Advantages of Our Tyrosine Analysis Service
- Universal dual-wavelength absorbance detector with higher sensitivity
- High data collection rate and accurate peak analysis
- Short analysis cycle and low price
- Using HPLC-MS platform to accurately determine the content of tyrosine
- Professional analysis engineer and provide one-stop service
Service Workflow
In our technical process, the sample does not need to undergo derivatization processing, and can be directly injected to achieve rapid and accurate determination of sample content. At the same time, it is less disturbed by baseline noise, making quantification more accurate and obtaining a more sensitive analysis report. LC-MS can detect and quantitative the content of trace sample substances.
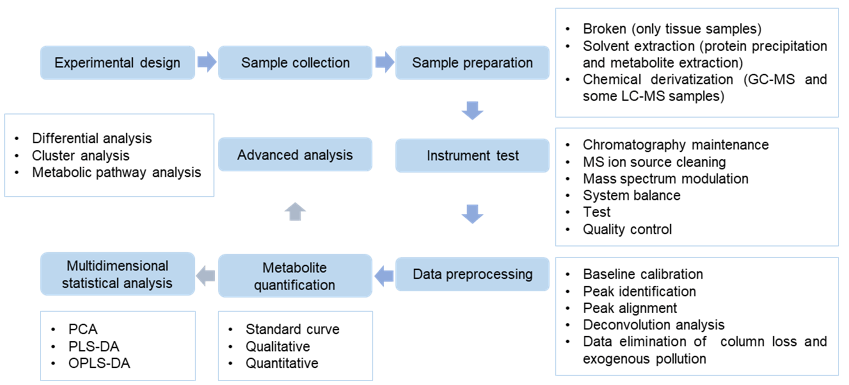 Figure 2. Tyrosine analysis service workflow.
Figure 2. Tyrosine analysis service workflow.
Detection method: HPLC-MS
Mobile phase: 8 mmol·L-1 potassium dihydrogen phosphate solution-methanol (9:1)
Injection volume: 10 μl
Flow rate: 2.0 ml/min
Elution mode: linear gradient elution
Detection wavelength: 280 nm
Resolution: 12.36-247.20 μg·ml-1
Precision: 0.58%
Average sample recovery rate: 99.4%
Repeatability: 89.5%
Reproducibility test: 86.1%
Detector: FLD
Analysis content:
- Standard straight line drawing
- Linear range and lower limit of quantification
- Stability and recovery rate test
- Chromatographic condition selection
- Chromatogram raw image and data collection
- Determination of tyrosine and its derivatives
Sample Preparation
- Serum/plasma: 50ul/sample
- Urine: 50µl/sample
- Tissue: 50mg/sample
- Fresh stool: 50mg/sample
- Freeze-dried stool: 5mg/sample
- Cell: 4×106/sample
Sample storage and transportation: Store in liquid nitrogen or -80°C, and transport on dry ice.
Number of biological replicates: At least 6 biological replicates for plant samples, and at least 10 biological replicates for animal samples
The samples include multiple sample types such as animals, plants and microorganisms, please contact us for details.
Delivery
- Complete experimental protocol
- Parameters for chromatograms and mass spectra
- Raw data files of chromatograms and mass spectra
- Analytical reports for the identification and content determination of Tyrosine and its derivatives
- Customized analytical reports
Creative Proteomics supports various types of amino acid and its metabolite analysis applications, which can help you quickly complete research and development work. If you would like to analyze other substances, please contact us.
References
- Bartesaghi S, Radi R. Fundamentals on the biochemistry of peroxynitrite and protein tyrosine nitration. Redox Biology. 2018;14:618-625.
- Cao M, Chen G, et al. Computational Prediction and Analysis for Tyrosine Post-Translational Modifications via Elastic Net. Journal of Chemical Information and Modeling. 2018;58(6):1272-1281.