Sinigrin (Allylglucosinolate) Analysis Service
Submit Your InquiryWhat is Sinigrin?
In the realm of plant chemistry, one compound that has been the focus of research is sinigrin, or allylglucosinolate, a type of glucosinolate compound found in various plants, particularly within the Brassica vegetable family. This secondary metabolite is generated as a means of defense against herbivores and other predators.
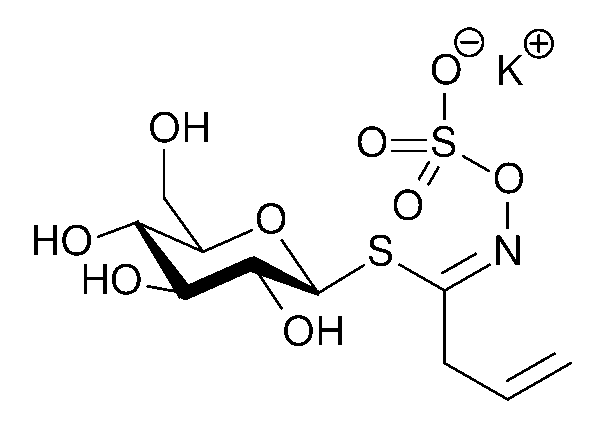 Structure of sinigrin
Structure of sinigrin
Sinigrin's potential health advantages have piqued the interest of researchers, as it exhibits anti-inflammatory, anti-cancer, and anti-oxidative properties. These therapeutic properties are believed to stem from the degradation of sinigrin into its biologically active derivatives, specifically allyl isothiocyanate and 1-cyano-2,3-epithiopropane.
Recent years have seen a rise in the importance of sinigrin analysis within different plant tissues and food products due to its potential health benefits. Such an analysis can offer valuable insight into the composition and qualities of this compound, as well as its potential applications in the food and pharmaceutical sectors.
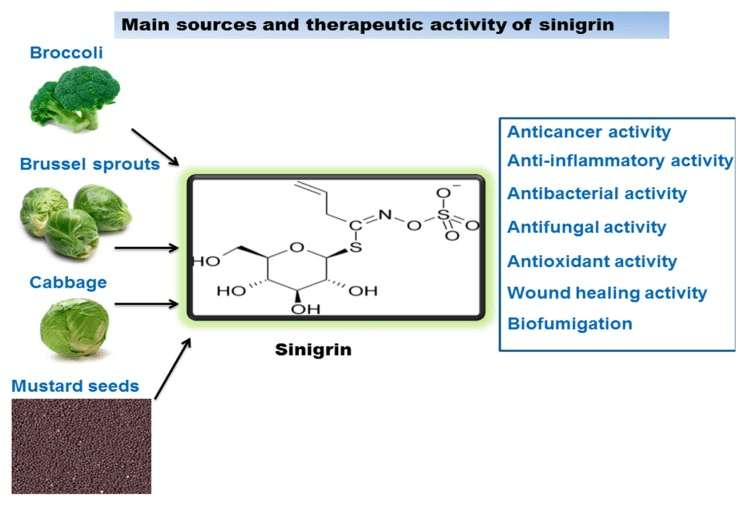 Sinigrin sources and its therapeutic activity (picture from Mazumder, Anisha, et al., 2016)
Sinigrin sources and its therapeutic activity (picture from Mazumder, Anisha, et al., 2016)
One of the most widely used methods for the analysis of sinigrin and its derivatives is liquid chromatography-mass spectrometry (LC-MS).
For LC-MS analysis of sinigrin, the sample is first extracted using a suitable solvent and then purified using solid phase extraction (SPE) or other techniques. The purified sample is injected into a liquid chromatography system where sinigrin and its derivatives are separated based on their chemical properties. The separated compounds are then detected and quantified using highly sensitive and selective mass spectrometry. LC-MS analysis of sinigrin can be performed using a variety of mass spectrometry techniques, including electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI). These techniques can provide accurate and precise quantification of sinigrin and its derivatives, even at low levels.
At Creative Proteomics, we offer LC-MS based sinigrin analysis services to meet the needs of our customers. Contact us to learn more about our sinigrin analysis services and how we can help advance your research.
Detection Platform for Sinigrin Analysis
- LC-MS/MS
- GC-MS
Waters ACQUITY UPLC, MS-SCIEX QTRAP 4500/5500/6500 and Waters Xevo TQ-S
Agilent 7890B-5977A, Thermo TRACE 1300-TSQ 9000
Technical Advantages
- High sensitivity
- Qualitative and quantitative standards
The sensitivity can reach pg level, and it can accurately quantify low-abundance metabolites.
Each metabolite is qualitatively and quantitatively compared with standard products to ensure the accuracy of the results.
Application Field
- Accurate quantification of allylglucosinolate (sinigrin)
- Validate hypotheses proposed by non-targeted metabolomics or other omics
- Research metabolic models for allylglucosinolate (sinigrin)
Technical Route of Sinigrin Analysis
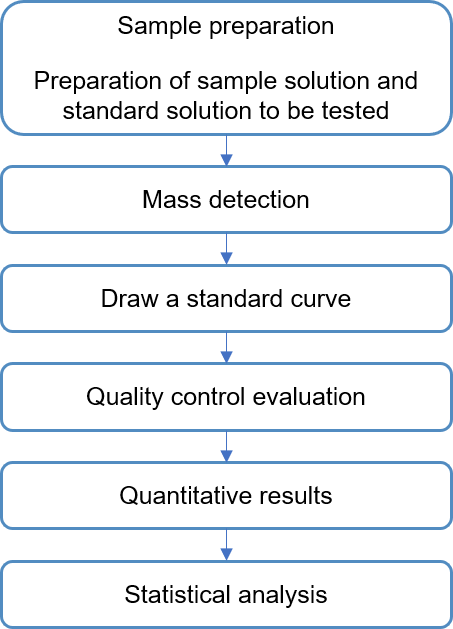
Sample Preparation Suggestions
| Sample type | Suggested sample quantity (per case) | Number of repetitions (recommended) |
|---|---|---|
| Plant tissue | ≥1g | ≥ 6 |
| Animals and clinical organizations | ≥200 mg | ≥ 10 |
| Serum, plasma | ≥200 mg | ≥ 6 |
| Urine | ≥200 μl | ≥ 6 |
| Stool, intestinal contents | ≥500 μl | ≥ 6 |
| Cell | ≥107 | ≥ 6 |
| Microorganism | ≥107 or ≥100 mg | ≥ 6 |
| Culture broth, fermentation broth | ≥1 ml | ≥ 6 |
Feedback to Customers
Creative Proteomics will provide you with detailed technical reports, including
- Experimental steps
- Related mass spectrometry parameters
- Part of the mass spectrum picture
- Raw data
- Quantitative results of targeted metabolites
Project Cycle
- A standard experimental procedure takes about 1 ~ 4 weeks.
Reference
- Mazumder, Anisha, et al. Sinigrin and Its Therapeutic Benefits. Molecules, 2016.