Inositol Phosphates Analysis Service
Submit Your InquiryInositol phosphates (IPs) are a group of mono to hexa-phosphorylated inositols. They play critical roles in diverse cellular functions including cell growth, apoptosis, cell migration, endocytosis and cell differentiation. Mono-, di-, and triphosphorylation of the inositol ring creates a wide range of stereochemically distinct signalling entities. For instance, inositol 1,4,5-trisphosphate (I(1,4,5)P3), is formed when the phosphoinositide phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) is hydrolysed by a phospholipase C isozyme. An array of inositol trisphosphate (IP3) and tetrakisphosphate (IP4) molecules are synthesised by the action of various kinases and phosphatases in the cytosol. These species then transport between the cytosol and the nucleus where they are acted on by inositol polyphosphate multikinase (IPMK), inositol-pentakisphosphate 2-kinase (IPPK), inositol hexakisphosphate kinase 1 (IP6K1) and 2 (IP6K2), to produce IP5, IP6, IP7, and IP8 molecules. As a carbohydrate metabolism, inositol phosphate metabolism plays important roles in mammalians. Thus, its dysfunction is closely associated with various diseases like Joubert syndrome and Centronuclear myopathy.
Inositol phosphates contain multiple phosphorylated derivatives and individual PIPs can contain different fatty acid chains, resulting in structural complexity. Creative Proteomics has built a dedicated analytical platform to provide you with qualitative and quantitative analysis of inositol phosphates. Based on ion chromatography separation techniques, we are able to accurately determine the total amount of phosphorylated derivatives. Based on LC/MS technology, we are able to quantify phosphatidylinositol containing specific fatty acid chain structures.
Advantages of Our Inositol Phosphates Analysis Service
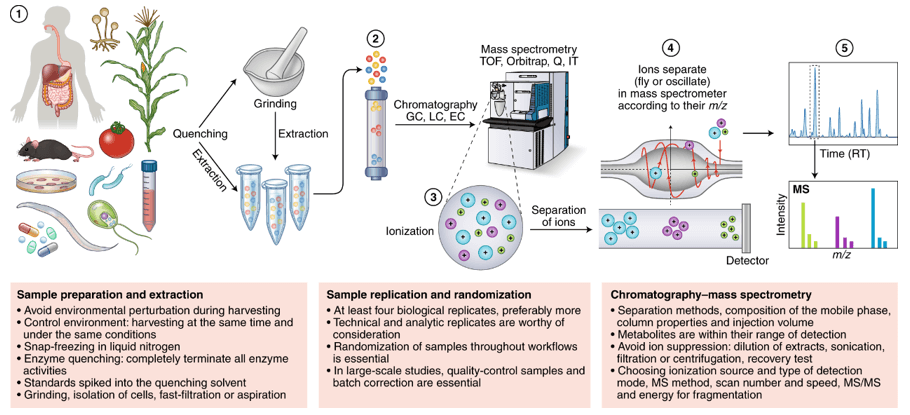
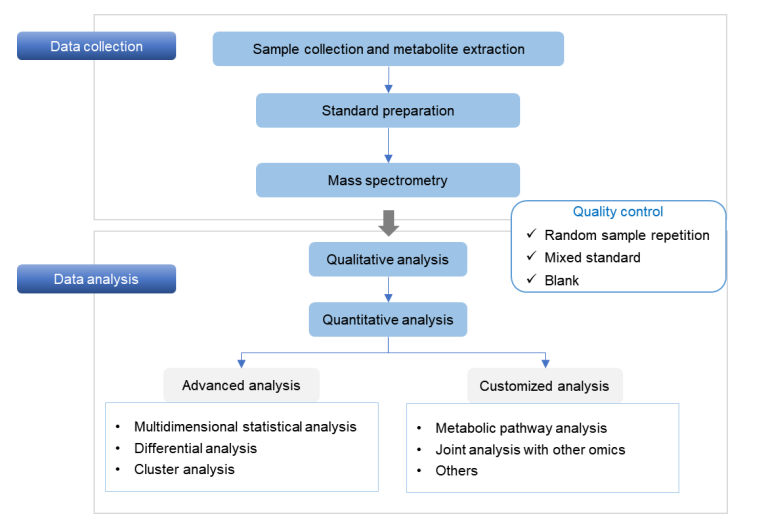
Service Workflow
Inositol Phosphates Related Analytes That We Can Measure
| Inositol monophosphate | Inositol bisphosphate | Inositol trisphosphate |
| Inositol pentakisphosphate | Inositol hexaphosphate | Myo-inositol |
| Bornesitol | Phosphatidylinositol-3,4,5-trisphosphate (PIP3) | Phosphatidylinositol 4,5-bisphosphate (PIP2) |
| 1-Phosphatidyl-D-myo-inositol | 1-Phosphatidyl-1D-myo-inositol 3-phosphate | D-myo-Inositol 1,3-bisphosphate |
| 1D-myo-Inositol 3-phosphate | 1D-myo-Inositol 1,3,4,5,6-pentakisphosphate |
Sample Requirements
We can analyze a wide range of biological materials including but not limited to cells and solid tissues from humans and animals, such as mice, rats, rabbits. If you need transport your samples to us, please follow the below requirements for different kinds of sample:
- Blood/plasma: 500ul/sample
- Urine: 1ml/sample
- Tissue: 200mg/sample
- Cells: 1x107/sample
- Feces: 500mg/sample
Shipment condition: dry ice
We can also accept other types of samples, please contact our technical staff for details.
Report Delivery
- Experimental procedures
- The parameters of liquid chromatograph and mass spectrometer
- HPLC and MS raw data files
- Metabolite quantitative data
Creative Proteomics has extensive experience in the analysis of substances. We will provide accurate and detailed data and analysis reports. If you would like additional information or to test other substances, you can contact us. We are looking forward to working with you.
References
- Monserrate Jessica P, York John D. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010,22:365-373.
- Bunney Tom D, Katan Matilda. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010.10:342-352.
- Bielas Stephanie L, Silhavy Jennifer L, Brancati Francesco, et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet. 2009,41:1032-1036.
- Agrawal Pankaj B, Pierson Christopher R, Joshi Mugdha, et al. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am J Hum Genet. 2014, 95:218-226.