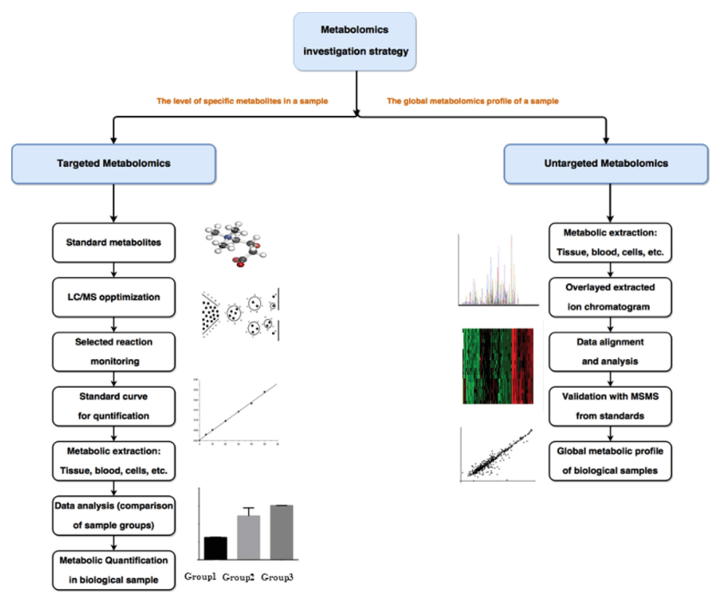
What Are Amino Sugars and Nucleotide Sugars—and Why Analyze Them?
Amino sugars and nucleotide sugars are essential precursors in glycan biosynthesis, glycoprotein formation, and bacterial cell wall assembly. They act as key substrates for glycosylation and metabolic signaling across biological systems.
However, their analysis is often hindered by poor chromatographic retention, co-elution of isomers, and instability during sample processing. These issues can compromise data quality in both discovery and quality control settings.
Creative Proteomics offers a targeted analysis service designed to overcome these challenges. We apply isomer-resolved separations, stabilization techniques, and sensitive LC–MS methods to support accurate, reproducible quantification. Our workflows are tailored for researchers investigating glycosylation pathways, metabolic regulation, or carbohydrate-based product consistency.
Targeted Services for Amino and Nucleotide Sugar Quantification
- Targeted quantification of free amino sugars
Includes glucosamine, galactosamine, mannosamine, and their N-acetylated forms (e.g., GlcNAc, GalNAc).
- Analysis of nucleotide-activated sugar donors
Covers UDP-sugars (e.g., UDP-Glc, UDP-GlcNAc), GDP-sugars (e.g., GDP-Fuc, GDP-Man), and CMP-sialic acids.
- Isomer-resolved sugar profiling
Methods designed to distinguish isobaric/isomeric pairs such as GlcNAc vs GalNAc, or Neu5Ac vs Neu5Gc.
- Stabilized detection of labile sugars
Customized derivatization and cold-chain sample handling to preserve sialic acids and phosphorylated sugars.
- Custom panel design and matrix validation
We support tailored compound panels and validate them in your specific sample type, including tissues, serum, microbial extracts, fermentation broths, and formulation buffers.
Full List of Detectable Amino Sugars and Nucleotide Sugars
Our targeted panels cover a comprehensive set of amino and nucleotide sugars relevant to glycosylation, bacterial cell wall synthesis, and sugar metabolism. Custom additions are also available upon request.
Amino Sugars
| Compound Name |
Abbreviation |
Notes |
| Glucosamine |
GlcN |
Common precursor in glycoproteins and GAGs |
| Galactosamine |
GalN |
Structural component of mucins and glycolipids |
| Mannosamine |
ManN |
Precursor for sialic acid biosynthesis |
| N-Acetylglucosamine |
GlcNAc |
Key substrate in N-glycosylation |
| N-Acetylgalactosamine |
GalNAc |
Involved in O-glycosylation initiation |
| N-Acetylmannosamine |
ManNAc |
Intermediate in sialic acid synthesis |
| Muramic acid |
Mur |
Unique to bacterial peptidoglycan |
| Sialic acids |
Neu5Ac, Neu5Gc |
Includes mono- and di-O-acetylated forms (selective detection) |
Nucleotide Sugars
| Compound Name |
Abbreviation |
Donor Type |
| UDP-Glucose |
UDP-Glc |
Hexose donor for glycan extension |
| UDP-Galactose |
UDP-Gal |
Used in β1-4 galactosylation |
| UDP-Glucuronic acid |
UDP-GlcA |
Precursor in proteoglycan and detox pathways |
| UDP-Xylose |
UDP-Xyl |
Essential for proteoglycan core structures |
| UDP-GlcNAc |
UDP-N-acetylglucosamine |
Central hub in glycan branching |
| UDP-GalNAc |
UDP-N-acetylgalactosamine |
O-glycosylation initiator |
| GDP-Mannose |
GDP-Man |
Core donor for high-mannose glycans |
| GDP-Fucose |
GDP-Fuc |
Fucosylation donor |
| GDP-Galactose |
GDP-Gal |
Less common but detectable |
| CMP-Neu5Ac |
CMP-sialic acid |
Donor for sialyltransferases |
| CMP-Neu5Gc |
CMP-sialic acid (variant) |
Less common in human cells |
| ADP-Glucose |
ADP-Glc |
Plant/microbial glycogen synthesis |
Not seeing your target? We routinely expand our method libraries. Contact us for compound evaluation and panel customization.
Why Choose Our Amino Sugar and Nucleotide Sugar Analysis Service?
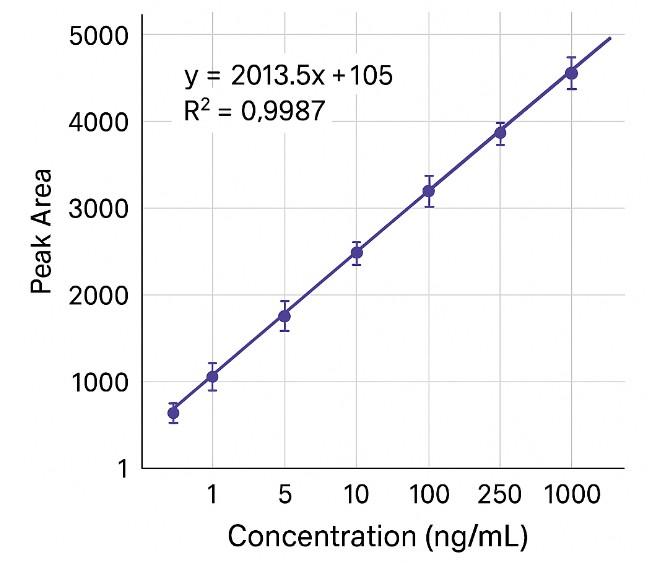
Typical limits of detection (LOD) range from <1 ng/mL to 5 ng/mL, depending on analyte chemistry and matrix complexity. Sensitivity can be enhanced through derivatization or enrichment when needed.
Calibration curves routinely achieve R2 ≥ 0.995 across dynamic ranges of two to three orders of magnitude, using matrix-matched standards or isotope-labeled internal controls.
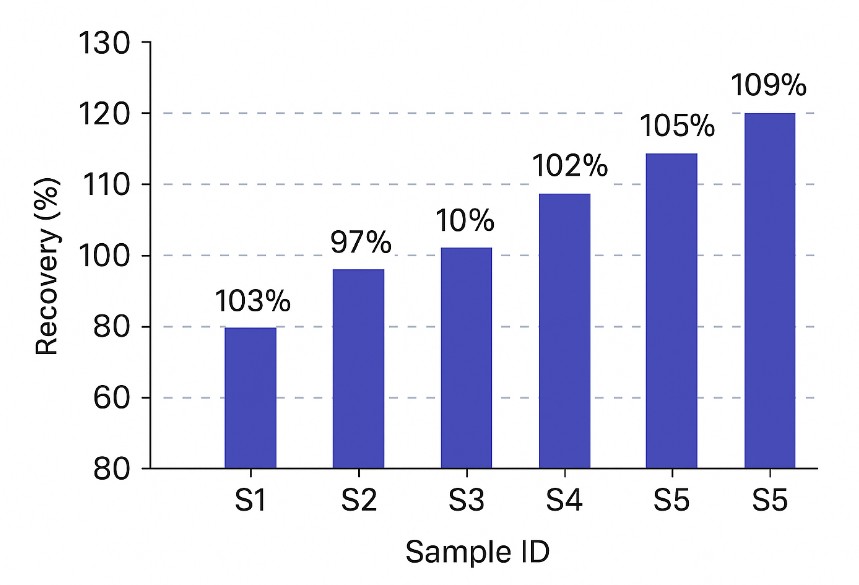
Inter- and intra-assay precision typically falls within 5–15% coefficient of variation (CV) for most analytes, based on triplicate injections and QC spike tracking.
High-resolution MS methods (Orbitrap or Q-TOF platforms) maintain <5 ppm mass error across batches, ensuring reliable compound identification even in isomer-rich panels.
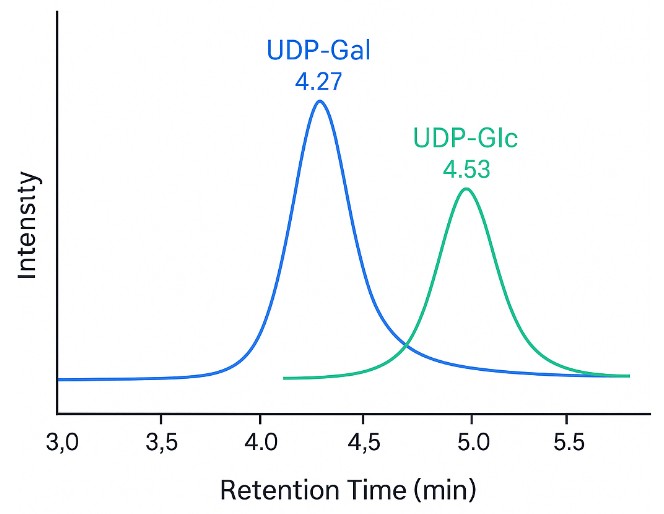
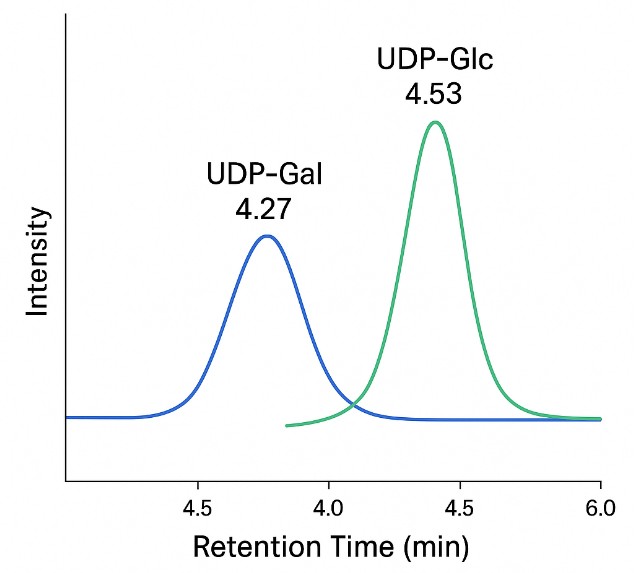
- Isomer-Specific Separation
HILIC and PGC-based LC methods achieve baseline separation of key isomers like GlcNAc vs GalNAc or Neu5Ac vs Neu5Gc—critical for pathway-specific interpretation.
- Validated Carryover Control
Optimized wash protocols and valve diversion steps reduce analyte carryover to <0.1% of the preceding peak area, protecting quantitation in large sample sets.
Every run includes blank injections, calibration standards, spike-in QCs, and system suitability tests to support transparent and auditable data.
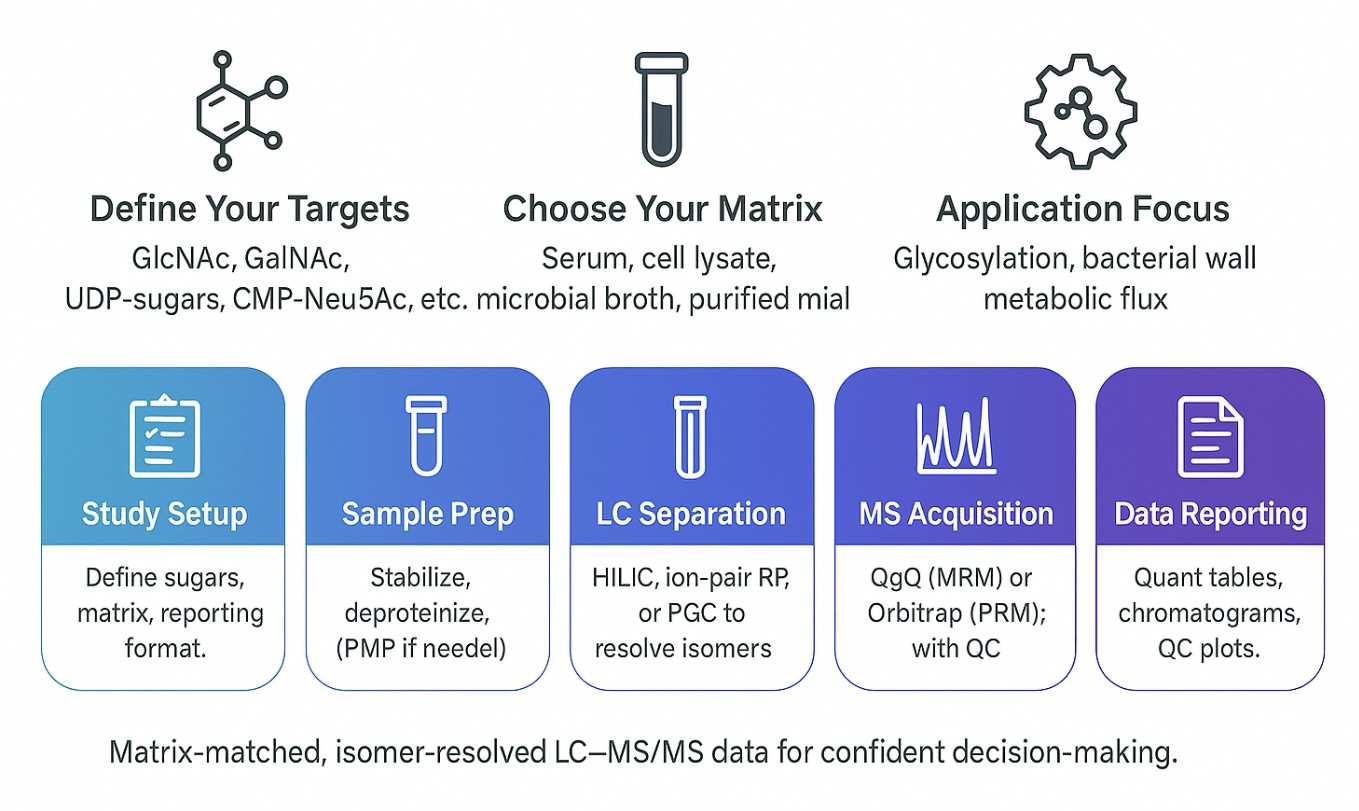
How We Analyze Amino and Nucleotide Sugars: Methods, Instruments, and Parameters
We use high-sensitivity UHPLC–MS/MS platforms with isomer-resolved separation to quantify amino sugars and nucleotide sugars in complex biological matrices. Methods are selected based on compound class, matrix complexity, and required sensitivity.
Instruments & Detection Modes
Orbitrap PRM / Full Scan (Q Exactive HF-X, Exploris 240): For high-resolution profiling with <5 ppm mass accuracy and 30k–60k resolving power.
Triple Quadrupole MRM (QTRAP 6500+, Xevo TQ-S): For targeted quantification with LOD <1–5 ng/mL in most matrices.
Separation Techniques
HILIC: Resolves amino sugars and N-acetyl derivatives (e.g., GlcNAc, GalNAc).
Ion-pair RP: Enhances retention of nucleotide sugars (e.g., UDP-, GDP-, CMP-linked).
PGC: Separates critical isomers like UDP-Glc vs UDP-Gal with baseline resolution.
Key Analytical Capabilities
| Capability |
What It Solves |
How It's Achieved |
| Isomer Resolution |
Prevents co-elution of GlcNAc vs GalNAc, UDP-Glc vs UDP-Gal |
HILIC or PGC columns + high-resolution MS |
| Labile Sugar Stability |
Avoids degradation of CMP-sialic acids, UDP-sugars |
pH control, rapid prep, derivatization (PMP/DMB) |
| Low-Level Detection |
Enables quantification in low-abundance samples |
LOD < 1–5 ng/mL via QqQ or Orbitrap PRM |
| Quantitative Accuracy |
Ensures reproducibility across batches and matrices |
Internal standards + matrix-matched calibration |
| Comprehensive QC |
Validates every data point |
Spike recovery, %CV, retention time checks |
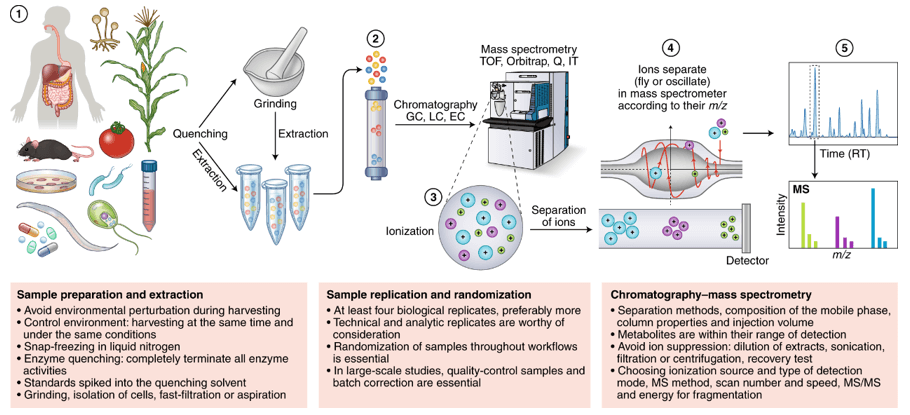
Step-by-Step Workflow for Amino and Nucleotide Sugar Quantification
Sample Requirements for Amino Sugar and Nucleotide Sugar LC–MS Analysis
| Sample Type |
Minimum Volume / Mass |
Container |
Notes |
| Biofluids (serum, plasma, urine, CSF, fermentation broth) |
≥100 µL |
Low-bind microcentrifuge tubes |
Avoid hemolysis; EDTA or heparin preferred |
| Tissues / Cells (animal, plant, microbial) |
≥20 mg tissue or cell pellet |
Cryovials or tubes |
Snap-freeze immediately after harvest; no non-volatile buffers |
| Lysates / Extracts |
≥100 µL |
Low-bind tubes |
Deproteinize if possible; record buffer composition |
| Purified Compounds / Intermediates |
≥50 µL or ≥2 mg |
Amber or low-adsorption vials |
Declare solvent and concentration; avoid surfactants or glycerol |
| Formulations / Media |
≥200 µL |
Sealed tubes or vials |
List all additives; high salt may require cleanup |
For matrices not listed above, please consult us before submission. Avoid using detergents, high-salt buffers, or preservatives unless necessary.
What You Receive: Deliverables from Our LC–MS/MS Sugar Analysis
- Quant tables: Analyte names, retention times, transitions or exact masses, concentration per sample, units, and QC flags.
- Calibration and QC pack: Calibration curves, residuals, R², back-calculated standards, spike-recovery, and precision metrics.
- Chromatographic evidence: Extracted ion chromatograms, peak annotations, and isomer-resolution notes.
- Method brief: Column, mobile phases, gradient outline, acquisition settings, internal standards, and sample prep summary.
- Raw data and method files: Vendor RAW/Wiff data, processing templates, and transition lists (on request).
- Pathway view (optional): Overlay of quantified metabolites on the amino sugar and nucleotide sugar pathway for interpretation.
Where Amino and Nucleotide Sugar Analysis Makes an Impact
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566