What is β-Oxidation Flux Analysis and Why It Matters
β-Oxidation flux analysis is a targeted metabolomics approach that quantifies the dynamic rate at which fatty acids are broken down via the β-oxidation pathway in mitochondria and peroxisomes. Instead of measuring only static metabolite concentrations, this method uses stable isotope tracers—such as [U-¹³C]-palmitate or [U-¹³C]-oleate—combined with high-sensitivity LC–MS/MS to track carbon flow through fatty acid activation, transport, and sequential oxidation cycles, ultimately generating acetyl-CoA for the tricarboxylic acid (TCA) cycle and downstream energy production.
In many research contexts, static lipid measurements cannot reveal pathway capacity, directionality, or bottlenecks. β-Oxidation flux analysis addresses this by providing quantitative, time-resolved insight into functional FAO activity, mitochondrial versus peroxisomal contributions, and metabolic rerouting under genetic, nutritional, or chemical perturbations. This is essential for understanding energy metabolism, optimizing bioprocess efficiency, validating mechanism-of-action studies, and guiding metabolic engineering strategies.
What Problems We Solve for Our Clients
Service Scope — Creative Proteomics β-Oxidation Flux Panel
- FAO Flux Quantification – Stable isotope tracing with ¹³C-labeled fatty acids, enrichment metrics, and optional model-based flux estimation.
- Acylcarnitine Profiling – Chain-length–resolved targeted profiling via LC–MS/MS with internal standard calibration.
- CoA Ester Quantitation – Quantification of key CoA esters using targeted MS methods with high accuracy and precision.
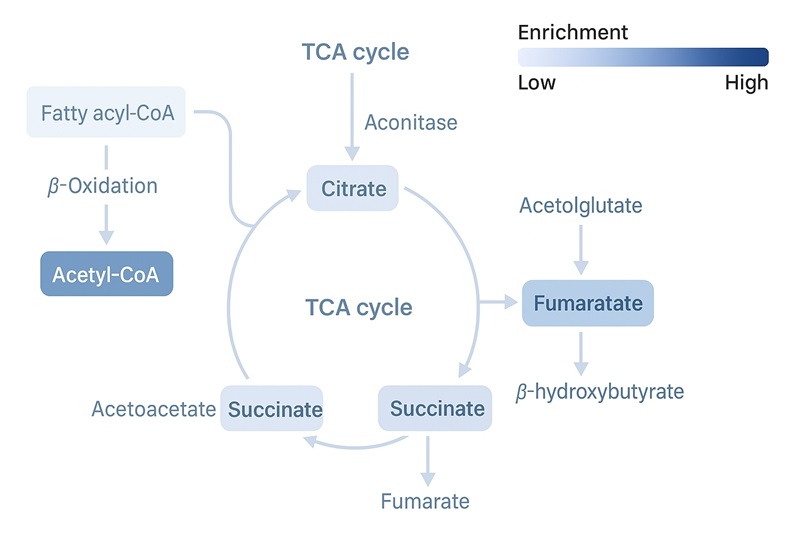
- TCA Isotopologue Mapping – Tracking carbon label incorporation into TCA intermediates to assess downstream metabolic flux.
- Mitochondrial vs Peroxisomal Contribution Assessment – Determine relative pathway activity using targeted acylcarnitine and dicarboxylic acid profiles. Tracer selection (e.g., chain length) can be tailored to emphasize mitochondrial or peroxisomal β-oxidation.
- Ketogenesis Readout – Measuring ketone body production and labeling to evaluate acetyl-CoA overflow.
- Multi-Tracer Substrate Competition Designs – Combining multiple labeled substrates to investigate fuel utilization and pathway interactions.
- Peroxisomal β-Oxidation Module – Extended coverage for very-long-chain fatty acid oxidation activity.
β-Oxidation Flux Panel – Analyte Coverage
Analyte classes covered in each module are detailed below:
| Metabolite Class |
Representative Analytes |
Notes |
| Free Carnitine and Short-Chain Acylcarnitines |
Free Carnitine (C0), Acetylcarnitine (C2), Propionylcarnitine (C3), Butyrylcarnitine (C4), Isobutyrylcarnitine (C4-i), Hydroxybutyrylcarnitine (C4-OH) |
Indicators of fatty acid activation and accumulation of initial β-oxidation products |
| Medium-Chain Acylcarnitines |
Hexanoylcarnitine (C6), Octanoylcarnitine (C8), Decanoylcarnitine (C10), Dodecanoylcarnitine (C12), Hydroxyoctanoylcarnitine (C8-OH), Hydroxydecanoylcarnitine (C10-OH) |
Assessment of medium-chain fatty acid oxidation efficiency and MCAD activity |
| Long-Chain Acylcarnitines |
Myristoylcarnitine (C14), Myristoleylcarnitine (C14:1), Hydroxymyristoylcarnitine (C14-OH), Palmitoylcarnitine (C16), Palmitoleylcarnitine (C16:1), Hydroxypalmitoylcarnitine (C16-OH), Stearoylcarnitine (C18), Oleoylcarnitine (C18:1), Linoleylcarnitine (C18:2) |
Reflect mitochondrial transport and degradation of long-chain fatty acids in β-oxidation |
| Very-Long-Chain Acylcarnitines (Peroxisomal Related) |
Tetracosanoylcarnitine (C24:0), Hexacosanoylcarnitine (C26:0), Tetracosenoic carnitine (C24:1) |
Evaluation of peroxisomal β-oxidation capacity and VLCFA metabolism |
| Dicarboxylic Acylcarnitines |
Glutarylcarnitine (C5-DC), Adipoylcarnitine (C6-DC), Suberylcarnitine (C8-DC), Sebacylcarnitine (C10-DC), Dodecanedioylcarnitine (C12-DC) |
Markers of ω-oxidation and compensatory metabolic pathway activity |
| CoA Esters |
Acetyl-CoA, Acetoacetyl-CoA, Succinyl-CoA, Malonyl-CoA, Palmitoyl-CoA |
Key intermediates linking fatty acid activation with TCA cycle entry |
| Ketone Bodies |
β-Hydroxybutyrate, Acetoacetate |
Indicators of acetyl-CoA overflow and ketogenesis |
| TCA Cycle Intermediates |
Citrate, α-Ketoglutarate, Succinate, Fumarate, Malate, Oxaloacetate (or derivatives) |
Track fatty acid–derived carbon flux into the TCA cycle |
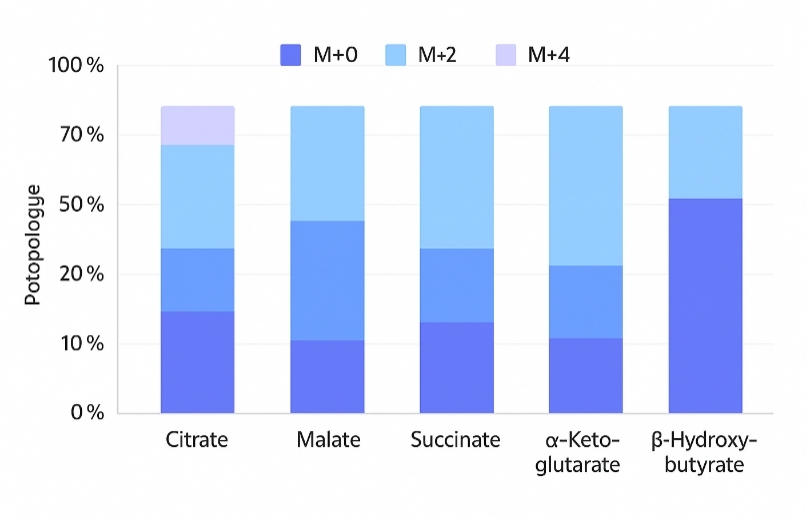
| Isotopologue Distributions |
Isotopologue patterns (M+1, M+2, …) for the above metabolites |
Quantify carbon flux and pathway utilization via ¹³C tracer incorporation |
Tracer Options and Study Modes
Primary ¹³C Tracers: [U-¹³C]-palmitate, [U-¹³C]-oleate, [1-¹³C]-palmitate.
Deuterated Standards: e.g., d₃-carnitine and chain-matched labeled acylcarnitines for accurate quantitation.
Static plus Flux Hybrid: Combine unlabeled quantitation with stable-isotope tracing for capacity and utilization in one design.
Multi-Tracer Designs: Use multiple labeled substrates to study fuel preference and metabolic rerouting.
Why Choose Our β-Oxidation Flux Panel: Key Advantages
- High Sensitivity Detection – Detects low nanomolar levels of acylcarnitines and organic acids.
- Excellent Quantitative Accuracy – Strong linearity with isotopically labeled internal standards.
- Reproducible Results – Consistent performance across batches.
- Comprehensive Analyte Coverage – Over 40 targeted species across short-, medium-, long-, and very-long-chain acylcarnitines, plus CoA esters, ketone bodies, and TCA intermediates.
- Dual-Platform Confidence – UHPLC–MRM quantitation plus optional HRMS confirmation.
- Isotopologue Precision – Reliable MID measurement with natural-abundance correction.
- Flexible Tracer Options – Multiple ¹³C-labeled fatty acids and multi-tracer designs.
Technical Specifications You Can Rely On
Platforms & Acquisition
- UHPLC–MS/MS on triple quadrupole (MRM) for targeted quantitation, with high-resolution MS (Orbitrap/Q-TOF) for confirmatory ID when requested.
- Dual-polarity switching with retention-time scheduling for maximal panel coverage.
- Calibration: Multi-point external calibration with isotopically labeled internal standards; typical linearity R² ≥ 0.995.
- Sensitivity: Method LOD/LOQ in the low nM to sub-µM range for acylcarnitines and many organic acids (matrix-dependent).
- Precision: Intra-/inter-batch %CV targets ≤ 15% for the majority of analytes in qualified matrices.
- Carryover Control: Assessed each batch with blanks and post-run rinses; acceptance criteria reported.
Flux Analytics
- MID extraction with isotope-correction; enrichment metrics presented with propagated error.
- Optional model-based flux estimation using established MFA tools (e.g., INCA/IsoCor-compatible outputs), with sensitivity analyses.
How Our β-Oxidation Flux Analysis Works — Step-by-Step Process
- Study Design Consulting – Define hypotheses, matrices, tracer(s), and readouts aligned to your decision points.
- Method Fit and Pilot (Optional) – Small-scale check of labeling performance, recovery, and analytical linearity in your matrix.
- Sample Preparation – Quench/extract under cold organic conditions; internal standards spiked at extraction.
- LC–MS/MS Acquisition – Scheduled MRM with matrix-matched calibration and pooled-QC monitoring.
- QA/QC Review – Blank evaluation, retention-time windows, peak shape, ion ratio confirmation, and batch-trend checks.
- Data Processing and Flux Calculations – MID generation, natural-abundance correction, enrichment and flux estimates.
- Reporting and Review – Delivery of results package and technical discussion focused on interpretation and next steps.
How to Prepare and Ship Samples for β-Oxidation Flux Studies
| Sample Type |
Minimum Amount |
Preparation |
Storage & Shipping |
| Cells |
≥ 1×10⁶ cells / condition |
Adherent or suspension; record culture medium composition for labeling |
Snap-freeze in liquid N₂; ship on dry ice |
| Tissues |
≥ 20–50 mg wet weight |
Remove excess blood/debris; snap-freeze immediately after collection |
Store at −80 °C; ship on dry ice |
| Biofluids |
≥ 50–100 µL plasma, serum, or conditioned media |
Centrifuge to remove particulates; aliquot to avoid freeze–thaw |
Ship on dry ice in leak-proof containers |
| Extracts (pre-prepared) |
Contact us for volume |
Cold organic extraction; add internal standards during extraction |
Store at −80 °C; ship on dry ice |
Note:
- Labeling Information: Provide tracer type, concentration, and labeling duration with the submission form.
- General Handling: Avoid repeated freeze–thaw cycles. Minimize processing time before freezing. Always ship on dry ice in secondary containment to prevent leakage.
Applications of β-Oxidation Flux Panel
Deliverables: What You Get from Our β-Oxidation Flux Analysis Service
Executive Summary: Key findings, pathway interpretation, and decision-oriented recommendations.
Quant Tables: Absolute concentrations (where applicable), normalized levels, and isotopologue matrices (.xlsx/.csv).
Flux Readouts: MID-based metrics (fractional enrichment, M+2/M+16 ratios where relevant) and model-derived flux estimates with confidence bounds.
Visual Package: Pathway maps annotated with enrichment, volcano/ratio plots for conditions, and QC dashboards.
Raw & Processed Data: Vendor files and open formats (.mzML/.raw as available), method metadata, and isotopic-correction reports.
Methods Appendix: Tracer chemistry, acquisition parameters, calibration models, and QA/QC acceptance criteria.
Macrophage-associated lipin-1 promotes β-oxidation in response to proresolving stimuli
Schilke, R. M., Blackburn, C. M., Rao, S., Krzywanski, D. M., Finck, B. N., & Woolard, M. D.
Journal: Immunohorizons
Year: 2020
Lipin-1 regulates lipid catabolism in pro-resolving macrophages
Schilke, R. M., Blackburn, C. M., Rao, S., Krzywanski, D. M., Finck, B. N., & Woolard, M. D.
Journal: bioRxiv
Year: 2020
Ketone bodies are mildly elevated in subjects with type 2 diabetes mellitus and are inversely associated with insulin resistance as measured by the lipoprotein insulin resistance index
Garcia, E., Shalaurova, I., Matyus, S. P., Oskardmay, D. N., Otvos, J. D., Dullaart, R. P., & Connelly, M. A.
Journal: Journal of Clinical Medicine
Year: 2020
Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism
Han, L., Huang, D., Wu, S., Liu, S., Wang, C., Sheng, Y., … & Yang, L.
Journal: Nature Cell Biology
Year: 2023
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024