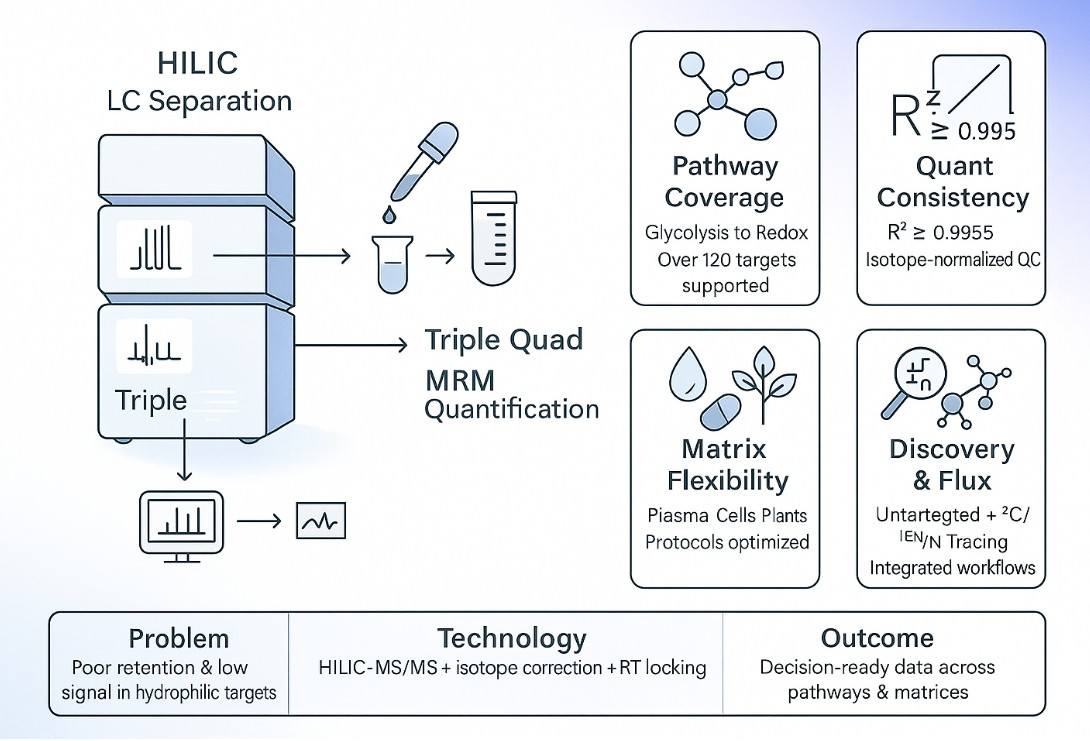
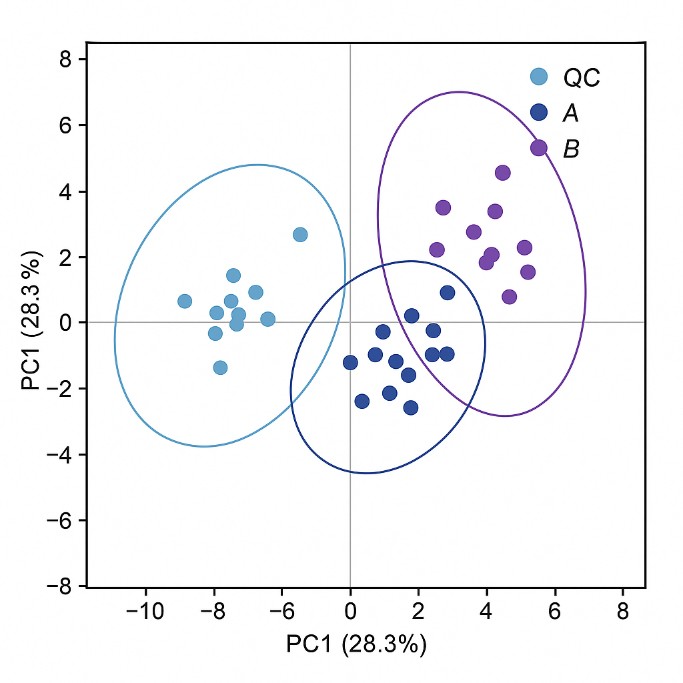
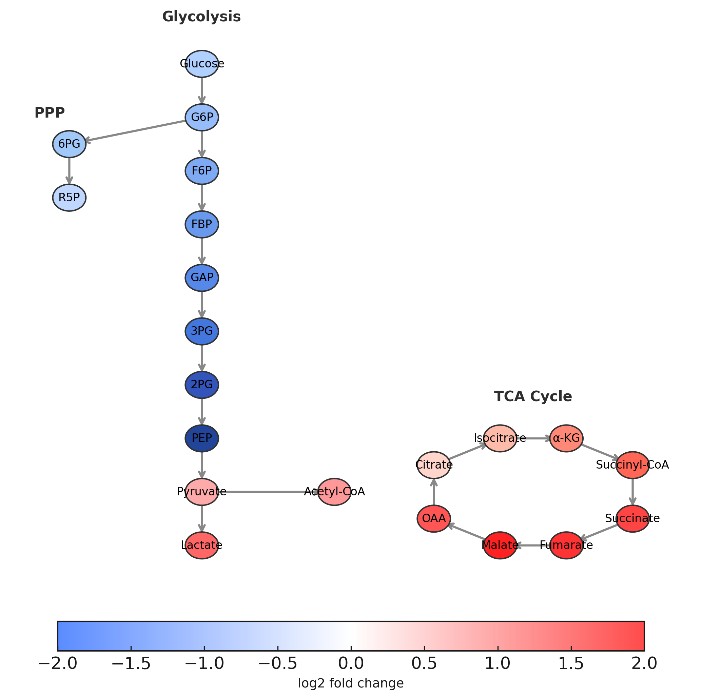
What Are Polar Metabolites and Why Analyze Them
Polar metabolites are small, water-soluble molecules that drive core biochemistry. They include amino acids, organic acids, nucleotides, and sugar phosphates. Together, they reflect glycolysis, the TCA cycle, and redox balance.
Polar metabolites analysis uncovers real-time pathway shifts under treatments or stress. It helps explain cellular energy use, biosynthetic demand, and cofactor status. These readouts support mechanism studies, bioprocess control, and quality assessment.
Because these compounds are highly hydrophilic, standard reversed-phase methods struggle. Dedicated workflows—such as HILIC-LC paired with mass spectrometry—improve retention, sensitivity, and coverage for confident decision-making.
Problems We Help You Solve
Creative Proteomics helps research teams overcome common challenges in polar metabolite analysis:
- Inconsistent quantification across complex biological matrices
- Low signal intensity for hydrophilic compounds in standard LC–MS workflows
- Missing data points due to poor retention or early elution
- Difficulty comparing studies due to batch effects and platform variability
- Unclear metabolite identities in untargeted datasets
- Limited panel coverage in pathway-specific studies
Our platform is designed to deliver high-confidence, decision-ready data even from challenging samples.
Our Polar Metabolite Analysis Capabilities
- Targeted Metabolite Quantification
Absolute or relative quantification of key polar metabolites using internal standards and calibration curves.
- Untargeted Polar Metabolomics
Broad-spectrum profiling to uncover metabolic shifts, differential features, or unknown biomarkers.
- Pathway-Specific Panels
Focused analysis of glycolysis, TCA cycle, pentose phosphate pathway, amino acid metabolism, or nucleotide turnover.
- Isotope-Labeled Metabolic Flux Analysis
13C/15N tracer experiments to track flux through central metabolic pathways under defined conditions.
- Matrix-Specific Method Development
Custom optimization for plasma, serum, urine, tissue, cell lysate, microbial broth, or plant material to ensure compatibility and reproducibility.
Full List of Detectable Polar Metabolites
Our platform supports broad, research-grade coverage of polar metabolites. The table lists representative compounds by pathway or class. It is not exhaustive. If your target is missing, contact us for a custom panel or method fit.
| Category |
Representative Analytes |
| Central Carbon-Glycolysis / Gluconeogenesis |
Glucose, Glucose-6-phosphate, Fructose-6-phosphate, Fructose-1,6-bisphosphate, Glyceraldehyde-3-phosphate, Dihydroxyacetone phosphate, 3-Phosphoglycerate, 2-Phosphoglycerate, Phosphoenolpyruvate, Pyruvate, Lactate |
| Pentose Phosphate Pathway (PPP) |
Ribose-5-phosphate, Ribulose-5-phosphate, Xylulose-5-phosphate, Sedoheptulose-7-phosphate, Erythrose-4-phosphate, 6-Phosphogluconate |
| TCA Cycle & Anaplerosis |
Citrate, Isocitrate, α-Ketoglutarate, Succinyl-CoA*, Succinate, Fumarate, Malate, Oxaloacetate, Acetyl-CoA*, Citraconate, Itaconate |
| Amino Acids & Derivatives |
Alanine, Arginine, Asparagine, Aspartate, Cysteine, Glutamate, Glutamine, Glycine, Histidine, Isoleucine, Leucine, Lysine, Methionine, Phenylalanine, Proline, Serine, Threonine, Tryptophan, Tyrosine, Valine, Hydroxyproline, GABA |
| Urea Cycle & Nitrogen Metabolism |
Ornithine, Citrulline, Argininosuccinate, Urea, Ammonium, Creatine, Creatinine |
| Nucleotides (Ribo/Deoxyribo) & Bases |
AMP/ADP/ATP, GMP/GDP/GTP, CMP/CDP/CTP, UMP/UDP/UTP, dAMP/dADP/dATP, dGMP/dGDP/dGTP, dCMP/dCDP/dCTP, dTMP/dTDP/dTTP, Inosine, Hypoxanthine, Xanthine, Adenosine, Guanosine, Cytidine, Uridine |
| Nucleotide Sugars |
UDP-Glucose, UDP-Galactose, UDP-N-acetylglucosamine (UDP-GlcNAc), UDP-N-acetylgalactosamine (UDP-GalNAc), UDP-Glucuronic acid, CDP-Choline, CMP-Neu5Ac |
| Coenzymes & Acyl-CoAs* |
Coenzyme A, Acetyl-CoA, Succinyl-CoA, Malonyl-CoA, Propionyl-CoA, Butyryl-CoA, Isobutyryl-CoA, Crotonyl-CoA |
| Redox Cofactors & Thiols |
NAD⁺/NADH, NADP⁺/NADPH, FAD, FMN, Glutathione (GSH/GSSG), Cystine, Cystathionine, Thioredoxin (peptide-level optional) |
| One-Carbon & Methylation |
SAM (S-adenosylmethionine), SAH, Homocysteine, Methionine, 5-Methyl-THF (coverage method-dependent), Choline, Betaine |
| Organic Acids (Broad) |
Acetate, Formate, Propionate, Butyrate, Glyoxylate, Tartrate, Malonate, Pyroglutamate, 3-Hydroxybutyrate |
| Osmolytes & Quaternary Amines |
Choline, Betaine, Carnitine, Acetylcarnitine, Trimethylamine N-oxide (TMAO), Taurine |
| Polyamines & Biogenic Amines |
Putrescine, Spermidine, Spermine, Agmatine, Ethanolamine |
| Sugar Alcohols & Polyols |
Inositol (myo-/scyllo-), Sorbitol, Mannitol, Glycerol |
| Sugar Derivatives |
N-Acetylglucosamine (free), N-Acetylgalactosamine (free), Glucosamine, Galactosamine |
| Vitamins & Water-Soluble Cofactors† |
Thiamine (B1), Riboflavin (B2), Niacinamide/Nicotinic acid (B3), Pantothenate (B5), Pyridoxal-5'-phosphate (B6), Biotin (B7), Folates†, Ascorbate |
| Miscellaneous Polar Compounds |
Uric acid, Allantoin, Inosine monophosphate (IMP), 3-Methylhistidine, Trimethyllysine |
* Acyl-CoA species require dedicated extraction and LC methods.
† Certain folates and labile vitamers may need targeted stabilization and protected handling.
Note: This is a partial, representative list. We routinely expand or customize panels for specific species, matrices, or pathways. If you do not see your target, contact us to discuss feasibility and method development.
Why Choose Our Polar Metabolite Analysis Service?
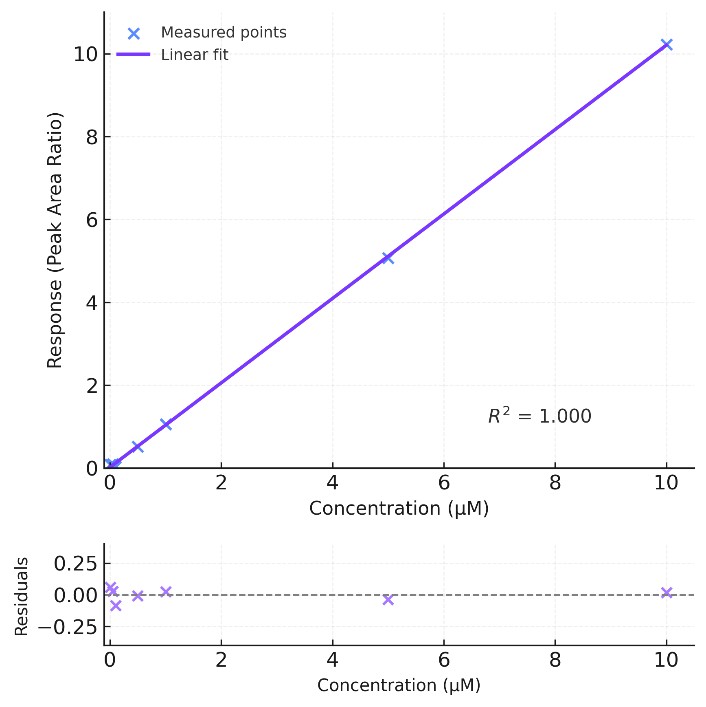
Multi-point calibration typically achieves R2 ≥ 0.995 across working ranges.
System suitability and pooled-QC controls keep inter-batch CV commonly ≤ 15%.
Optimized HILIC–MS/MS methods reach femtomole to low-picomole on-column limits.
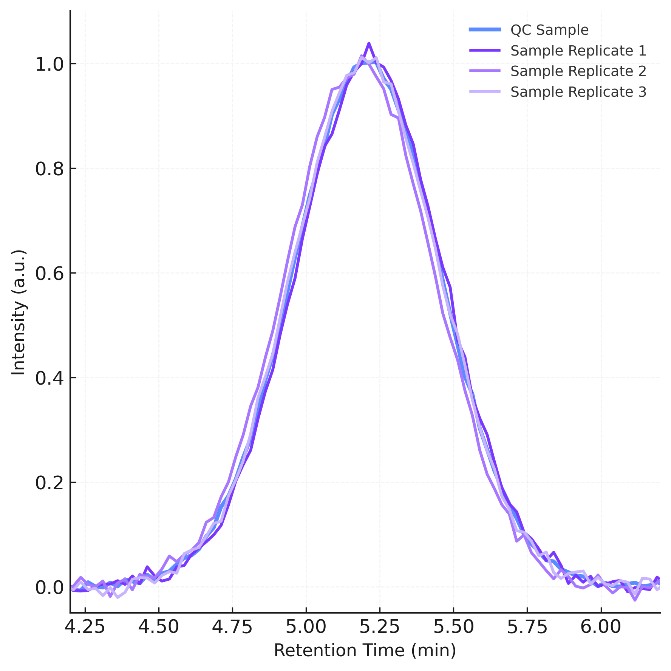
RT windows locked with internal standards reduce mismatches and carryover risk.
Stable-isotope spikes correct matrix effects for more reliable concentrations.
Drift correction and RT alignment improve cross-study comparability and trend analysis.
Curated panels span central carbon, nucleotides, sugar phosphates, and thiol/redox species.
LC–MS/MS Platforms and Detection Parameters for Polar Metabolites
Our polar metabolite analysis service is built on a robust LC–MS/MS framework designed to handle the chemical diversity and hydrophilicity of low-abundance metabolites. All analyses are conducted under strict quality controls to ensure accuracy, sensitivity, and reproducibility across sample types.
We primarily use hydrophilic interaction liquid chromatography (HILIC) coupled with:
Orbitrap Exploris 480
High-resolution mass spectrometry (HRMS) for untargeted profiling and metabolite identification.
- Mass accuracy: < 2 ppm
- Resolution: up to 480,000 FWHM
SCIEX Triple Quad™ 6500 Plus
Targeted MRM-based quantification with high sensitivity and low background noise.
- Dynamic range: > 105
- Inter-batch RSD: ≤ 15% (QC-controlled)
Separation is optimized using 2.1 × 100 mm HILIC columns (1.7 µm), with temperature control (±0.1 °C) to stabilize retention times. Internal standards and pooled QC samples are included in every batch to support multi-point calibration (R2 ≥ 0.995) and accurate cross-sample comparisons.
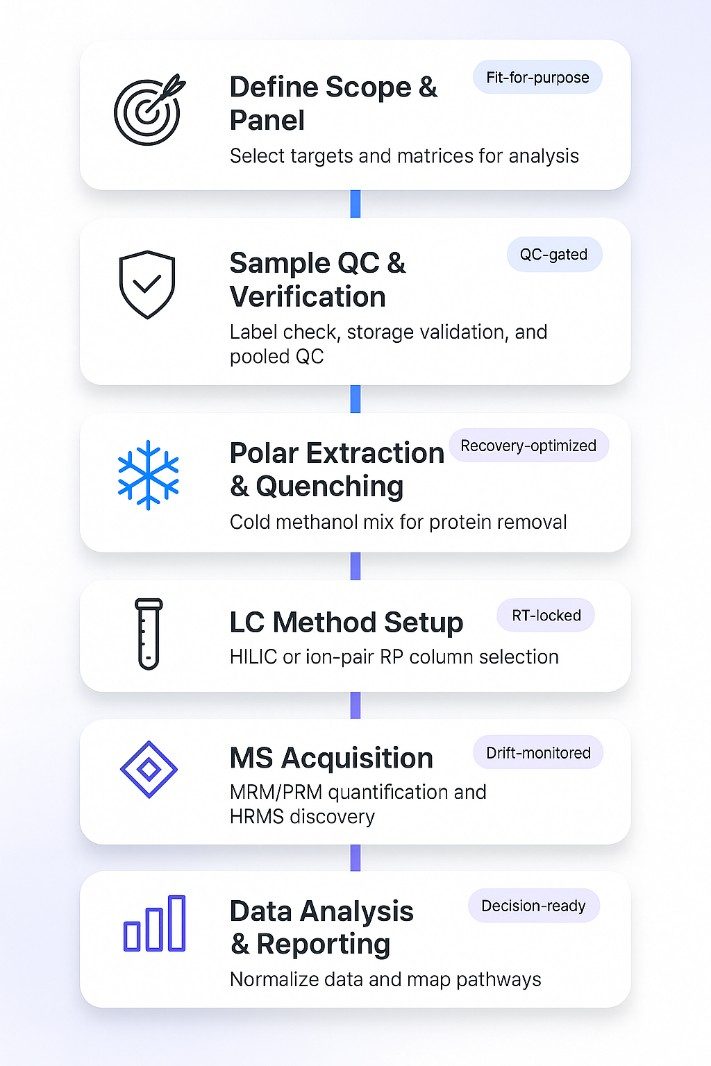
Polar Metabolite Analysis Workflow: Step by Step
Sample Requirements for Polar Metabolites Profiling
The following guidelines help protect labile polar compounds and ensure high-quality data. If your matrix differs or input is limited, contact us for a fit-for-purpose plan.
| Sample Type |
Minimum Amount |
Container |
Pretreatment |
Storage |
Shipping |
| Plasma / Serum |
100 µL |
Screw-cap cryovial |
Fast separation from cells; avoid hemolysis |
−80 °C |
Dry ice |
| Whole Blood |
300 µL |
EDTA tube + cryovial |
Immediate cold quench, spin ≤10 min |
−80 °C |
Dry ice |
| Urine |
500 µL |
Screw-cap cryovial |
Mix well; avoid preservatives |
−80 °C |
Dry ice |
| Tissue (mammalian) |
50–100 mg |
Pre-cooled cryotube |
Snap-freeze in liquid N2; no buffer |
−80 °C |
Dry ice |
| Cell Pellet |
≥1×106 cells |
Cryovial |
Wash in cold PBS; remove supernatant fully |
−80 °C |
Dry ice |
| Microbial Pellet |
Wet pellet from 10–20 mL culture |
Cryovial |
Rapid chill; remove media thoroughly |
−80 °C |
Dry ice |
| Plant Material |
100–200 mg |
Pre-cooled cryotube |
Flash-freeze; avoid water rinses |
−80 °C |
Dry ice |
| Culture Supernatant / Media |
1–2 mL |
Screw-cap tube |
Clarify by cold spin; no additives |
−80 °C |
Dry ice |
| Extracts (if self-prepared) |
100–200 µL |
LC-compatible vial |
80% MeOH/H2O, cold; no salts/detergents |
−80 °C |
Dry ice |
What You Receive: Deliverables from Our Polar Metabolite Analysis
- Raw LC–MS/MS files (.raw/.wiff) with run logs
- Quant tables (peak areas, concentrations) with internal-standard normalization, calibration fit, QC flags (CSV/XLSX)
- ID evidence (retention time, precursor/fragment ions, mass error, reference match notes)
- QC summary (pooled-QC trends, RT stability, carryover checks, batch diagnostics)
- Figures (annotated chromatograms, MS/MS spectra, PCA/volcano plots, pathway overlays)
- Methods brief (column, mobile phase, key MS parameters, processing settings)
- README & data dictionary (file map, variable definitions, units)
Applications of Polar Metabolites Analysis in Research and Industry
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566