What Are Thiols—and Why Analyze Them
Thiols are small sulfur-containing molecules featuring a reactive sulfhydryl (–SH) group. As key participants in redox reactions, enzyme catalysis, and cellular detoxification, they serve as sensitive indicators of oxidative balance, metabolic flux, and thiol–disulfide exchange dynamics across diverse biological and chemical systems.
Precise thiol analysis is essential for understanding redox pathways, assessing sample integrity, and identifying unwanted chemical reactivity in biological, industrial, or formulation matrices. Yet due to their high reactivity and poor stability, thiols are among the most technically challenging metabolites to measure—requiring specialized derivatization and stabilization protocols to ensure data accuracy.
Problems We Help You Solve
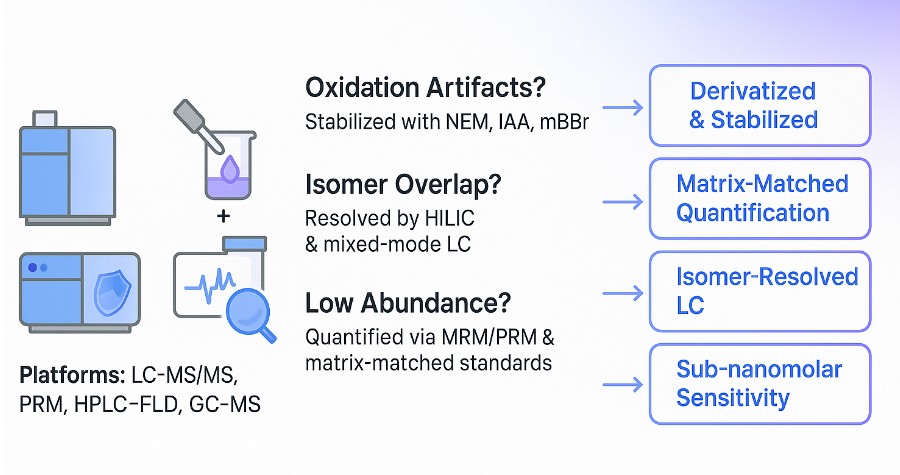
Thiols are chemically unstable and analytically difficult to quantify. Common issues include:
- Oxidation during handling
Free thiols readily oxidize to disulfides, distorting true concentrations unless rapidly stabilized.
- Poor retention and co-elution
Many thiols are small and polar, making them hard to retain on standard LC columns and prone to matrix interference.
- Derivatization bias
Incomplete or inconsistent labeling can compromise both sensitivity and accuracy.
Creative Proteomics overcomes these barriers using tailored stabilization, isomer-resolved chromatography, and matrix-matched quantification. The result is reliable, reproducible data—even from complex or reactive samples.
Thiols Analysis Service Options and Custom Modules
- Targeted LC–MS/MS Quantification
Quantitative MRM/PRM for predefined thiols panels across biofluids, cells, tissues, and process matrices; fit-for-purpose calibration and QC.
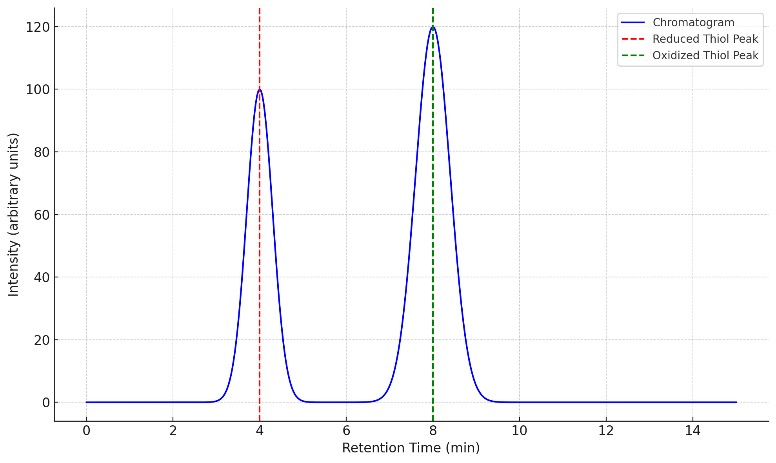
- Redox Speciation (Reduced vs Disulfide Forms)
Stabilized workflows to measure free, protein-bound, and oxidized states; supports redox ratio readouts and trend tracking.
- High-Resolution Confirmation (PRM/HRAM)
Orthogonal confirmation of key targets or suspected adducts; documentation suitable for method transfer.
- Volatile Thiols (GC–MS)
Headspace or extract-based methods for aroma-/process-related sulfur compounds in food, beverage, and fermentation contexts.
- Custom Method Development or Bridging
Method transfer, matrix-effect studies, spike-recovery optimization, and SOP alignment for regulated-like environments.
Detectable Thiols and Sulfur Compounds: Full Panel
| Category |
Representative analytes |
Notes |
| Core endogenous low-MW thiols |
Cysteine (free/protein-bound), Homocysteine, Cysteinylglycine, Cysteamine, γ-Glutamylcysteine, 2-Mercaptoethanol |
Speciation of free vs. protein-bound available |
| Disulfides & redox pairs |
Cystine, Homocystine, Mixed disulfides (Cys–GSH, GSH–protein), GSSG |
Report with redox-pair context if needed |
| RSNO & persulfides (reactive sulfur species) |
S-Nitrosoglutathione (GSNO*), S-Nitrosocysteine (Cys-NO*), Glutathione persulfide (GSSH*), Cysteine persulfide (Cys-SSH*) |
*Stabilized capture/derivatization required |
| Sulfenic/sulfinic/sulfonic acids (oxidized thiol states) |
Cysteine-SOH*, Cysteine-SO2H, Cysteic acid (Cys-SO3H) |
*Trapping reagents recommended for SOH |
| Glutathione pathway & conjugates |
GSH, GSSG, Ophthalmate, Selected GSH-conjugates (custom list) |
Targeted MRM/PRM with reference standards |
| CoA thiols & acyl-thioesters |
CoA-SH, Acetyl-CoA, Succinyl-CoA, Malonyl-CoA, Propionyl-CoA, Butyryl-CoA, Palmitoyl-CoA, Crotonyl-CoA |
Panel extendable to additional acyl-CoAs |
| Sulfur metabolites (inorganic/related) |
H2S*, Thiosulfate, Sulfite/Sulfate†, Taurine, Cysteinesulfinic acid |
*Captured/derivatized; †reported as related anions |
| Volatile thiols (aroma/process) |
3-Mercaptohexan-1-ol (3-MH), 3-Mercaptohexyl acetate (3-MHA), 2-Furfurylthiol, Methanethiol, Ethanethiol, Benzenemethanethiol, 3-Mercapto-3-methyl-1-butanol |
GC–MS headspace or extract workflows |
| Exogenous/therapeutic thiols & reagents |
N-Acetylcysteine (NAC), Penicillamine, DTT†, β-Mercaptoethanol†, MESNA (2-mercaptoethanesulfonate), Thiocholine |
†Typically tracked as process controls |
| Thioethers & special sulfur antioxidants |
Ergothioneine (ERG), Lanthionine |
Often quantified alongside thiols for redox context |
| Cyclized/derived forms |
Homocysteine thiolactone (HCTL), Cystathionine |
Include when homocysteine metabolism is in scope |
Notes: Final panels are tailored to matrix and purpose. If a target is not listed, provide a standard or structure and we will scope feasibility.
Why Choose Our Thiols Analysis Service?
Predefined quench/derivatization plans reduce oxidation artifacts and re-equilibration errors.
Method acceptance targets: R2 ≥ 0.995 and ≤ 15% CV intra-batch, documented per batch.
- Matrix-Matched Recovery (85–115%):
Calibration is performed in matched matrices (serum, broth, buffers) to minimize suppression; spike recovery data is reported per batch.
Calibration routinely spans ≥ 4 orders of magnitude, with matrix-matched verification.
- Trace detection when matrices permit
Triple-quadrupole MRM supports sub-nanomolar detection; achieved LOD/LOQ reported by analyte.
- Isomer-resolved chromatography
HILIC or mixed-mode methods minimize co-elution and ion suppression for polar thiols.
- Instrument-level traceability
Delivered packages include raw data, method files, transitions, and calibration back-calculations.
Sequence design and hardware rinses target < 0.1% carryover; blanks confirm performance.
PRM/HRAM verification flags interferences and secures identity in complex matrices.
How We Analyze Thiols: Methods, Instruments, and Parameters
At Creative Proteomics, thiols are quantified using targeted LC–MS/MS, PRM-based high-resolution confirmation, or HPLC-FLD, depending on analyte class, matrix type, and project goals. All methods are derivatization-compatible and stability-aware.
LC–MS/MS (Targeted Quantification)
Platforms:
- Triple Quad: Agilent 6495C
- UHPLC Systems: Waters ACQUITY
Method Highlights:
- Scheduled MRM for throughput and matrix-specific transitions
- HILIC or mixed-mode chromatography for polar thiols and redox pairs
- Derivatization reagents include NEM, IAA, mBBr
Best suited for: Absolute quantification in plasma, tissues, cells, fermentation samples
HPLC–FLD (Fluorescence Detection)
Platform: Agilent 1260 Infinity II FLD
Reagents: Monobromobimane (mBBr) or Thioglo tags
Application: Routine free thiol quantification or GSH:GSSG ratio tracking
Best suited for: High-throughput sample screening without isomer resolution
High-Resolution PRM (Qualitative & Orthogonal Confirmation)
Platform: Orbitrap Exploris 480
Use Case: Identity confirmation, interference resolution, conjugate screening
Approach: PRM with full MS2 spectra, optional library matching or neutral loss scanning
Best suited for: Adduct verification, low-abundance confirmation, unknown annotation
GC–MS (Volatile Sulfur Compounds)
Platform: Thermo ISQ 7000
Approach: Headspace-SPME, derivatization or direct injection
Analytes: Methanethiol, ethanethiol, 3-MH, 2-furfurylthiol
Best suited for: Food aroma studies, process volatiles, fermentation diagnostics
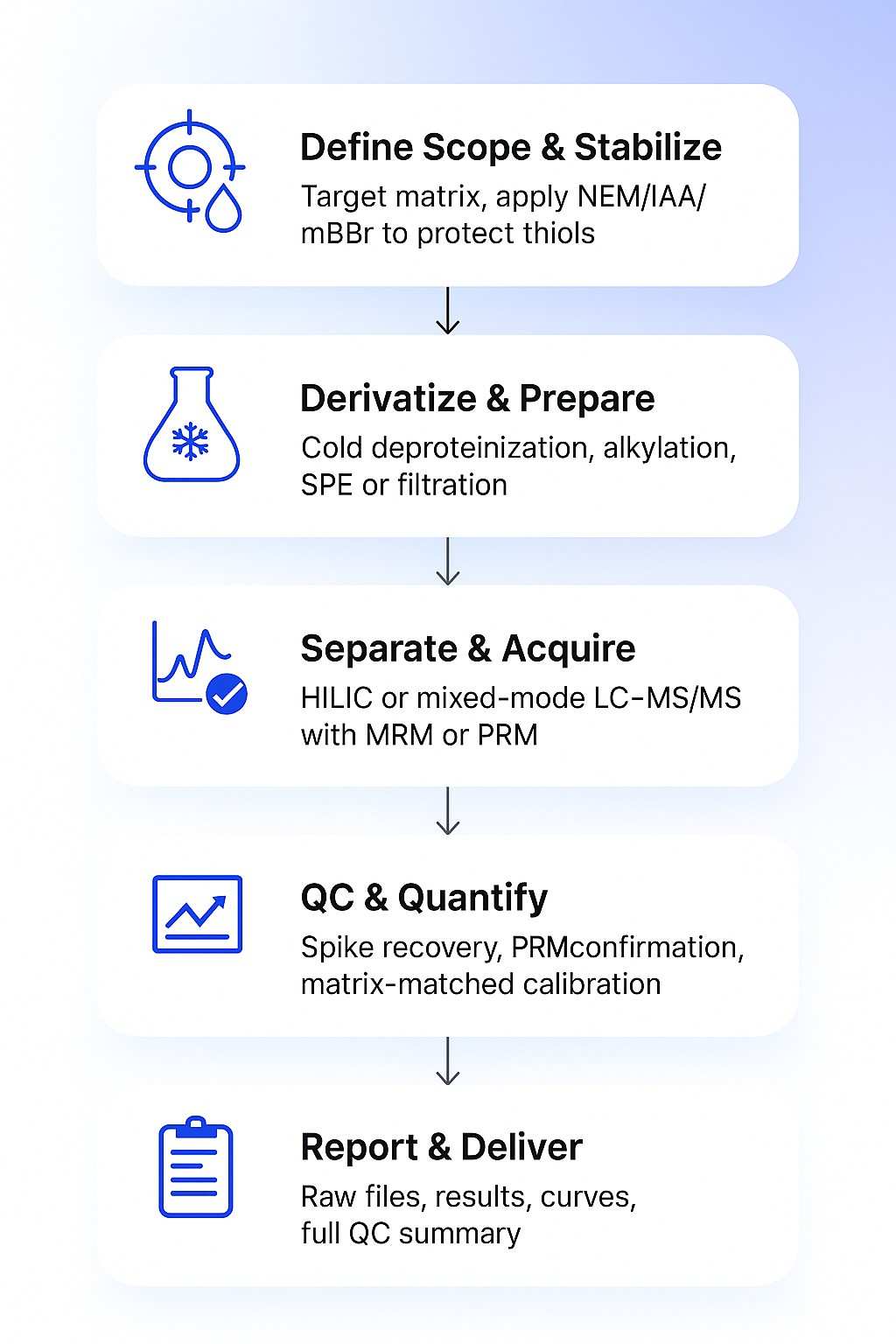
Thiols Analysis Workflow: Step by Step
Sample Requirements for Thiols Profiling
| Sample Type |
Minimum Volume |
Recommended Concentration |
Storage Conditions |
Preparation Notes |
| Biological Fluids |
100 µL |
1–5 mg/mL |
Store at -80°C for long-term storage |
Ensure samples are collected in clean, sterile containers to avoid contamination. Use EDTA or other anticoagulants when necessary. |
| Tissue Samples |
50 mg |
10–100 mg/g |
Store at -80°C or snap freeze with liquid nitrogen for long-term storage |
Samples should be processed immediately after collection to prevent oxidation. Homogenize tissue samples in buffer containing protease inhibitors. |
| Cell Culture Media |
200 µL |
0.1–10 mg/mL |
Refrigerate at 4°C for short-term or store at -80°C for long-term |
Collect media at the appropriate time point to capture relevant thiol changes. Filter to remove cell debris before analysis. |
| Protein Solutions |
10 µL |
1–10 mg/mL |
Store at -80°C for long-term storage |
Ensure protein samples are stored in small aliquots to prevent repeated freeze-thaw cycles, which can affect thiol stability. |
| Plasma/Serum |
100 µL |
1–5 mg/mL |
Store at -80°C or freeze immediately |
Plasma or serum should be separated from whole blood within 1–2 hours of collection to prevent oxidation and protein degradation. |
| Plant Samples |
100 mg |
10–100 mg/g |
Store at -80°C for long-term storage |
Homogenize plant material with a suitable extraction buffer immediately after collection to prevent oxidation of thiol groups. |
Notes for All Sample Types:
- Avoid exposure to light during sample collection and processing, as thiols are sensitive to UV radiation.
- Use antioxidants in buffers or solutions to minimize thiol oxidation during sample preparation.
- Minimize sample handling to avoid degradation or modification of thiol groups.
What You Receive: Deliverables from Our Thiols Analysis
- Analytical Report: A detailed summary of thiol concentrations, oxidation states, and disulfide bond analysis.
- Raw Data Files: Raw data in CSV, Excel, or instrument-specific formats for further analysis.
- Statistical Analysis: Interpretation of the data, including statistical significance and comparisons.
- Data Visualizations: Graphs and charts for easy interpretation of results.
- Methodology Document: A description of the methods and protocols used in the analysis.
Applications of Thiols Analysis in Research and Industry
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., Khalid, K., Van Le, H., Teo, H. M., Raitelaitis, M., ... & Prabhu, N.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
High Levels of Oxidative Stress Early after HSCT Are Associated with Later Adverse Outcomes
Cook, E., Langenberg, L., Luebbering, N., Ibrahimova, A., Sabulski, A., Lake, K. E., ... & Davies, S. M.
Journal:Transplantation and Cellular Therapy
Year: 2024
DOI: https://doi.org/10.1016/j.jtct.2023.12.096
Multiomics of a rice population identifies genes and genomic regions that bestow low glycemic index and high protein content
Badoni, S., Pasion-Uy, E. A., Kor, S., Kim, S. R., Tiozon Jr, R. N., Misra, G., ... & Sreenivasulu, N.
Journal: Proceedings of the National Academy of Sciences
Year: 2024
DOI: https://doi.org/10.1073/pnas.2410598121
The Brain Metabolome Is Modified by Obesity in a Sex-Dependent Manner
Norman, J. E., Milenkovic, D., Nuthikattu, S., & Villablanca, A. C.
Journal: International Journal of Molecular Sciences
Year: 2024
DOI: https://doi.org/10.3390/ijms25063475
UDP-Glucose/P2Y14 Receptor Signaling Exacerbates Neuronal Apoptosis After Subarachnoid Hemorrhage in Rats
Kanamaru, H., Zhu, S., Dong, S., Takemoto, Y., Huang, L., Sherchan, P., ... & Zhang, J. H.
Journal: Stroke
Year: 2024
DOI: https://doi.org/10.1161/STROKEAHA.123.044422
Pan-lysyl oxidase inhibition disrupts fibroinflammatory tumor stroma, rendering cholangiocarcinoma susceptible to chemotherapy
Burchard, P. R., Ruffolo, L. I., Ullman, N. A., Dale, B. S., Dave, Y. A., Hilty, B. K., ... & Hernandez-Alejandro, R.
Journal: Hepatology Communications
Year: 2024
DOI: https://doi.org/10.1097/HC9.0000000000000502
Comparative metabolite profiling of salt sensitive Oryza sativa and the halophytic wild rice Oryza coarctata under salt stress
Tamanna, N., Mojumder, A., Azim, T., Iqbal, M. I., Alam, M. N. U., Rahman, A., & Seraj, Z. I.
Journal: Plant‐Environment Interactions
Year: 2024
DOI: https://doi.org/10.1002/pei3.10155
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., Tseng, L., Mambetsariev, B., Li, H., Lou, C. H., Pozhitkov, A., ... & Salgia, R.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Physiological, transcriptomic and metabolomic insights of three extremophyte woody species living in the multi-stress environment of the Atacama Desert
Gajardo, H. A., Morales, M., Larama, G., Luengo-Escobar, A., López, D., Machado, M., ... & Bravo, L. A.
Journal: Planta
Year: 2024
DOI: https://doi.org/10.1007/s00425-024-04484-1
A personalized probabilistic approach to ovarian cancer diagnostics
Ban, D., Housley, S. N., Matyunina, L. V., McDonald, L. D., Bae-Jump, V. L., Benigno, B. B., ... & McDonald, J. F.
Journal: Gynecologic Oncology
Year: 2024
DOI: https://doi.org/10.1016/j.ygyno.2023.12.030
Glucocorticoid-induced osteoporosis is prevented by dietary prune in female mice
Chargo, N. J., Neugebauer, K., Guzior, D. V., Quinn, R. A., Parameswaran, N., & McCabe, L. R.
Journal: Frontiers in Cell and Developmental Biology
Year: 2024
DOI: https://doi.org/10.3389/fcell.2023.1324649
Proteolytic activation of fatty acid synthase signals pan-stress resolution
Wei, H., Weaver, Y. M., Yang, C., Zhang, Y., Hu, G., Karner, C. M., ... & Weaver, B. P.
Journal: Nature Metabolism
Year: 2024
DOI: https://doi.org/10.1038/s42255-023-00939-z
Quantifying forms and functions of intestinal bile acid pools in mice
Sudo, K., Delmas-Eliason, A., Soucy, S., Barrack, K. E., Liu, J., Balasubramanian, A., … & Sundrud, M. S.
Journal: bioRxiv
Year: 2024
DOI: https://doi.org/10.1101/2024.02.16.580658
Elevated SLC7A2 expression is associated with an abnormal neuroinflammatory response and nitrosative stress in Huntington's disease
Gaudet, I. D., Xu, H., Gordon, E., Cannestro, G. A., Lu, M. L., & Wei, J.
Journal: Journal of Neuroinflammation
Year: 2024
DOI: https://doi.org/10.1186/s12974-024-03038-2
Thermotolerance capabilities, blood metabolomics, and mammary gland hemodynamics and transcriptomic profiles of slick-haired Holstein cattle during mid lactation in Puerto Rico
Contreras-Correa, Z. E., Sánchez-Rodríguez, H. L., Arick II, M. A., Muñiz-Colón, G., & Lemley, C. O.
Journal: Journal of Dairy Science
Year: 2024
DOI: https://doi.org/10.3168/jds.2023-23878
Glycine supplementation can partially restore oxidative stress-associated glutathione deficiency in ageing cats
Ruparell, A., Alexander, J. E., Eyre, R., Carvell-Miller, L., Leung, Y. B., Evans, S. J., ... & Watson, P.
Journal: British Journal of Nutrition
Year: 2024
DOI: https://doi.org/10.1017/S0007114524000370
Untargeted metabolomics reveal sex-specific and non-specific redox-modulating metabolites in kidneys following binge drinking
Rafferty, D., de Carvalho, L. M., Sutter, M., Heneghan, K., Nelson, V., Leitner, M., ... & Puthanveetil, P.
Journal: Redox Experimental Medicine
Year: 2023
DOI: https://doi.org/10.1530/REM-23-0005
Sex modifies the impact of type 2 diabetes mellitus on the murine whole brain metabolome
Norman, J. E., Nuthikattu, S., Milenkovic, D., & Villablanca, A. C.
Journal: Metabolites
Year: 2023
DOI: https://doi.org/10.3390/metabo13091012
A human iPSC-derived hepatocyte screen identifies compounds that inhibit production of Apolipoprotein B
Liu, J. T., Doueiry, C., Jiang, Y. L., Blaszkiewicz, J., Lamprecht, M. P., Heslop, J. A., ... & Duncan, S. A.
Journal: Communications Biology
Year: 2023
DOI: https://doi.org/10.1038/s42003-023-04739-9
Methyl donor supplementation reduces phospho‐Tau, Fyn and demethylated protein phosphatase 2A levels and mitigates learning and motor deficits in a mouse model of tauopathy
van Hummel, A., Taleski, G., Sontag, J. M., Feiten, A. F., Ke, Y. D., Ittner, L. M., & Sontag, E.
Journal: Neuropathology and Applied Neurobiology
Year: 2023
DOI: https://doi.org/10.1111/nan.12931
Sex hormones, sex chromosomes, and microbiota: identification of Akkermansia muciniphila as an estrogen-responsive bacterium
Sakamuri, A., Bardhan, P., Tummala, R., Mauvais-Jarvis, F., Yang, T., Joe, B., & Ogola, B. O.
Journal: Microbiota and Host
Year: 2023
DOI: https://doi.org/10.1530/MAH-23-0010
Living in extreme environments: a photosynthetic and desiccation stress tolerance trade-off story, but not for everyone
Gajardo, H. A., Morales, M., López, D., Luengo-Escobar, M., Machado, A., Nunes-Nesi, A., ... & Bravo, L.
Journal: Authorea Preprints
Year: 2023
DOI: https://doi.org/10.22541/au.168311184.42382633/v2
Resting natural killer cell homeostasis relies on tryptophan/NAD+ metabolism and HIF‐1α
Pelletier, A., Nelius, E., Fan, Z., Khatchatourova, E., Alvarado‐Diaz, A., He, J., ... & Stockmann, C.
Journal: EMBO Reports
Year: 2023
DOI: https://doi.org/10.15252/embr.202256156
Function and regulation of a steroidogenic CYP450 enzyme in the mitochondrion of Toxoplasma gondii
Asady, B., Sampels, V., Romano, J. D., Levitskaya, J., Lige, B., Khare, P., ... & Coppens, I.
Journal: PLoS Pathogens
Year: 2023
DOI: https://doi.org/10.1371/journal.ppat.1011566