Glycosylphosphatidylinositol Anchor Analysis Service
Submit Your InquiryOverview
The GPI anchor is a complex glycophospholipids, and the C-terminus attached to the secreted protein after post-translated. GPI anchor precursor (CP2: complete precursor 2), accumulating in GPI transamidase mutants, including four mannose (Man1, Man2, Man3 and Man4), three ethanolamine phosphate (ETN-P) substituent on Man1, Man 2 and Man3, an acyl phosphatidylinositol (Acyl-Pi) and an amino glucose (GLCN). More than 20 genes encoding GPI anchored biological synthesis have been identified by genetic screening to isolate mutant cells of GPI proteins. The GPI anchor begins with the cytoplasmic side of the ER membrane. The first step in GPI biosynthesis is to add N-acetamin glucose (GLCNAc) to phosphatidylinositol (PI). The transfer reaction was executed through GPI-GlcNAc transferase, which was catalyzed by GPI15, ERI1, GPI3, GPI19, GPI2, and GPI1. Next, the acetyl group of GLCNAc was removed by Gpi12, a GlcNAc-PI de-N-acetylase and GlcN-PI was generated, which was transferred to the luminal side of the ER membrane.
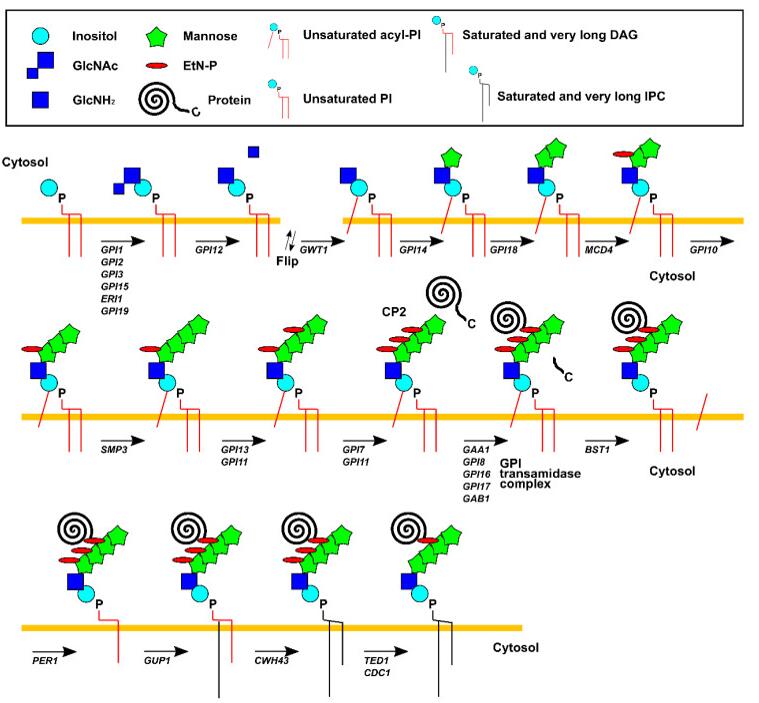 Fig 1. Pathways of GPI biosynthesis in Saccharomyces cerevisiae.
Fig 1. Pathways of GPI biosynthesis in Saccharomyces cerevisiae.
Inside the ER, the acyltransferase Gwt1 adds an acyl chain, which is mostly palmitate, to the 2-position of the inositol in GlcN-PI, generating GlcN-(acyl) PI. Subsequently, Man1 and Man2 were are transferred from dolicholphosphomannose (Dol-P-Man) to GlcN-(acyl) PI by GPI mannosyl-transferase 1 (Gpi14) and GPI mannosyl-transferase 2 (Gpi18), respectively. Further, ETN-P was transferred from phosphatidylethanolamine (PE) to Man1 by ETN-P transferase (MCD4). Then, Man 3 is added to the Man2 by GPI mannosyl-transferase 3 (GPI10), and the Man4 is added to Man3 through SMP3 to add ETN-P to MAN3 before the addition of EtN-P to Man3 by EtN-P transferase 3. The complex contains Gpi13 as a catalytic subunit and a stabilizing subunit Gpi11. Finally, EtN-P was added by adding EtN-P transferase 2, which consists of the catalytic subunit GPI7 and the stabilizing subunit Gpi11.
Advantages
- Short time-consuming
- High sensitivity and few detection restrictions
- High precision and good repeatability
- High throughput
- Customized service
Service workflow
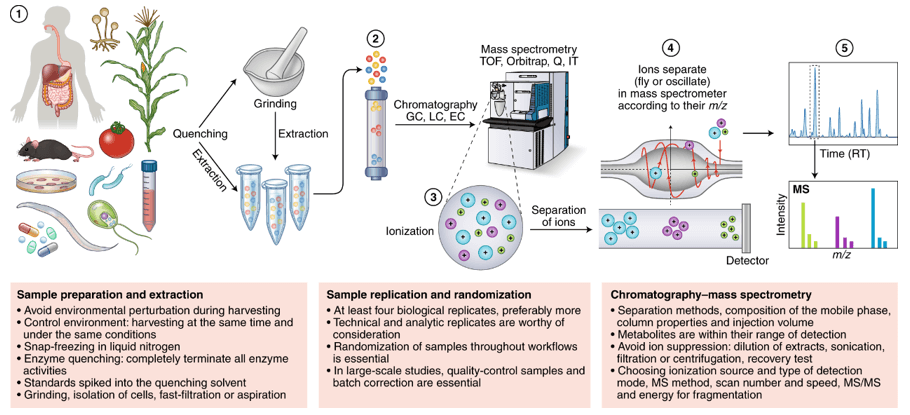
Lipid rafts are microscopic regions enriched in GPI-APs cholesterol, sphingolipids, transmembrane proteins, and lipidated proteins. The phospholipid of the GPI anchor is essential for the hydrophobicity of GPI-AP. By treatment with phosphatidylinositol specific phospholipase C(PI-PLC) or phospholipase D(PLD), a GPI anchor can be cleaved between the phosphate and lipid. When temperature-induced phase separation was performed in Triton X-114, a hydrophilic form of GPI-APs cleaved by PI-PLC or PLD was found in aqueous phase. GPI-Aps, PI-PLC and PLD isolated from Arabidopsis thaliana treat human lipid raft-associated proteins. This method be able to effectively indicate the presence of GPI-AP and use mass spectrometry to identify the ω-site.
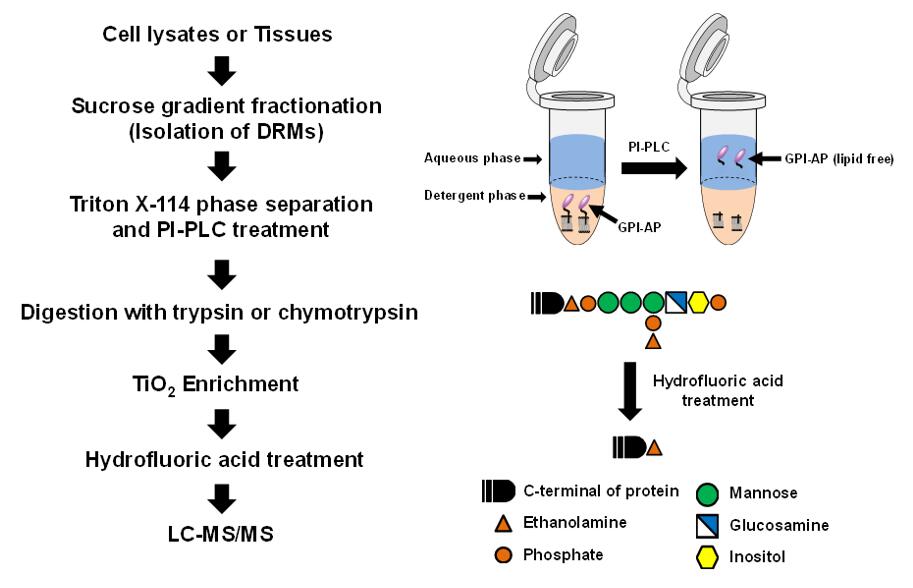 Fig 2. Work flow of GPI analysis based on LC-MS/MS.
Fig 2. Work flow of GPI analysis based on LC-MS/MS.
Sample requirement
- Serum/plasma: 500 μl/sample
- Protein: 100 µg
- Anticoagulated blood (EDTA): 1 mL
- Urine: 1 ml/sample
- Animal tissues: 200 mg/sample
- Cells: ≥ 1 × 107/sample
- Feces: 500 mg/sample
Repeated freezing and thawing of samples must be avoided. The serum sample should be precipitated in the collection tube for 30 minutes at room temperature, then transported to the centrifuge tube and centrifuged at 8000 rpm for 5 minutes. After centrifugation, the supernatant was equally divided into a freezing tube of 500 uL / sample.
Anticoagulants and preservatives must be added immediately after collection and then frozen at -80 °C.
Urine samples should be equally divided into centrifuge tubes with 1 mL per tube, each tube is added with 1/100 (w/v) sodium azide and stored at -80 °C.
Samples should be frozen in liquid nitrogen immediately and then transported to -80 °C for storage after collected.
Cytoactive should be terminated immediately to maintaining cell integrity.
In general, to assure enough sample to fulfill the whole project, the volume of the single sample need to be offered as much as possible. The remaining samples will be stored for one year free of charge and returned at any time if necessary. All samples need to be stored and transported at -80°C and try to avoid using surfactants (SDS, Triton-X) and inorganic salts.
Clinical samples are repeated in no less than 30 cases in a single group.
Animal samples are repeated in no less than 9 cases in a single group.
Report delivery
- Experimental procedure
- Parameters of HPLC and MS
- Raw data, chromatograms and mass spectra
- Metabolites quantification data
- Custom analysis report
Service cycle
- Sample testing: 5-10 working days
- Data analysis: 5-10 working days
Creative Proteomics metabolism analysis platform is committed to the all-around, reliable and accurate analysis service for a variety of target substances, which is suitable for life-science research, drug exploration, biological determination and other fields. We sincerely hope to cooperate with you to assistant your scientific research.
References
- W. Wang, L. Shi, Y. Qin, et al. Research and Application of Chondroitin Sulfate/Dermatan Sulfate-Degrading Enzymes. Front Cell Dev Biol, 2020, 8:560442.
- Z. Wang, K. Arnold, Y. Xu, et al. Quantitative analysis of heparan sulfate using isotopically labeled calibrants. Commun Biol, 2020, 3(1):425.