Cyclic nucleotides—cAMP, cGMP, and cGAMP—are essential second messengers that convert extracellular cues into precise intracellular responses. However, due to their extremely low endogenous levels and rapid enzymatic turnover, they remain difficult to quantify with conventional metabolomics methods.
Creative Proteomics offers a specialized LC-MS/MS platform optimized for the absolute quantification of these labile molecules, delivering the sensitivity, specificity, and reproducibility required for drug development, signaling studies, and mechanism-of-action research.
What Problems This Service Helps You Solve
Measuring cyclic nucleotides requires more than just high-end hardware; it requires a specialized approach to biology and chemistry. Our service helps you solve:
- The Sensitivity Gap: Detecting femtomole-level concentrations in limited samples like primary cells or tumor biopsies.
- Rapid Enzymatic Turnover: Preventing the immediate degradation of cAMP/cGMP by phosphodiesterases (PDEs) during sample collection.
- Isomer Ambiguity: Specifically distinguishing between signaling isomers like 2'3'-cGAMP (mammalian) and 3'3'-cGAMP (bacterial) that ELISA cannot resolve.
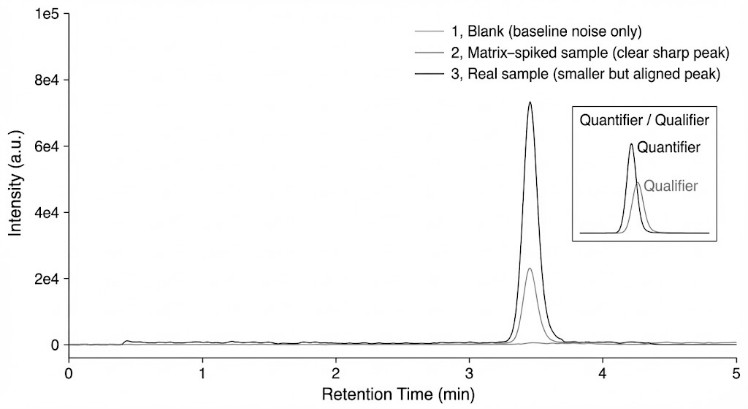
- Matrix Suppression: Eliminating interference from high-salt or complex biological backgrounds using optimized HILIC separation.
Service Options And Analyte Coverage For cAMP, cGMP, And cGAMP
Choose a focused single-analyte readout, or add pathway context with a panel. Optional variants and add-ons are available when they improve interpretation.
What We Can Quantify
| Scope |
Included Analytes |
When To Choose |
| Single Analyte |
cAMP or cGMP or cGAMP |
One messenger is your primary decision metric |
| Core Panel |
cAMP + cGMP + cGAMP |
You want signaling context under one QC plan |
| Optional Variants (Project-Dependent) |
2'3'-cGAMP, 3'3'-cGAMP |
When isomer specificity matters (standards/matrix dependent) |
| Related Nucleotides (Add-On) |
AMP, GMP, ATP, GTP (and others as needed) |
Upstream/downstream interpretation (project-defined) |
Built-In Quality Options
- Standard QC: calibration + QC samples + blanks/carryover checks (fit-for-purpose)
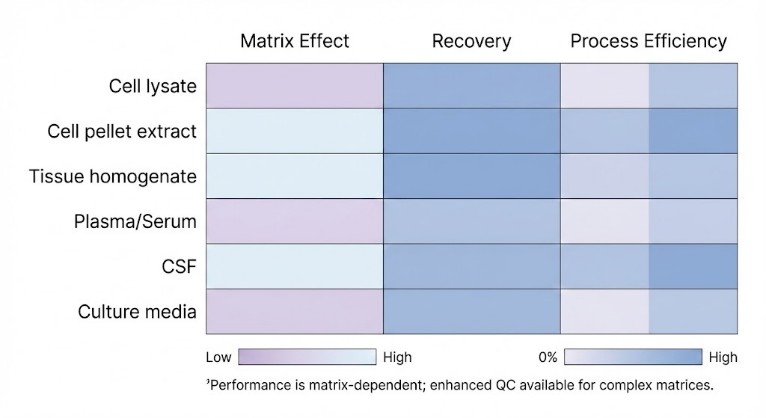
- Enhanced QC for challenging matrices: spike/recovery and matrix-effect checks when backgrounds vary widely
Quick Decision Guide: Single-Analyte Vs. Panel
| Your Research Goal |
Recommended Scope |
Why This Works |
Common Enhancements |
| Verify whether one messenger changes |
Single analyte |
Fastest, most focused interpretation |
Absolute quantification + Standard QC |
| Interpret signaling direction or balance |
Panel |
Context reduces misinterpretation |
Project-defined ratio-style summaries |
| STING pathway engagement |
cGAMP ± Panel |
Targets pathway messenger directly |
Isomer-aware support; Enhanced QC |
| Multiple matrices / variable backgrounds |
Panel + Enhanced QC |
Improves comparability across sample types |
Spike/recovery; matrix-effect checks |
Advantages Of Targeted LC-MS/MS For Second Messengers
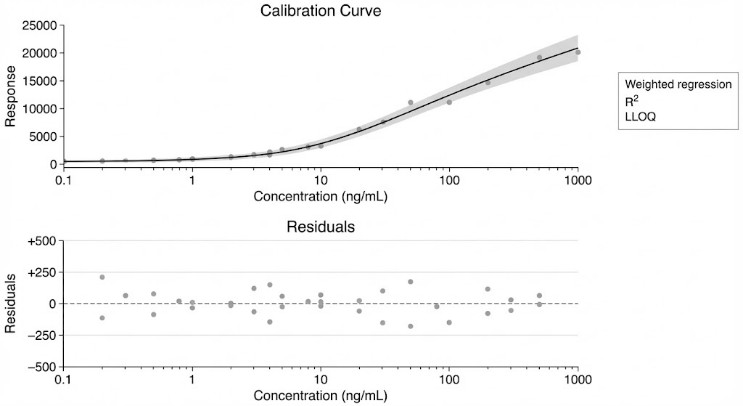
- Sensitivity (Project-Dependent): LLOQ can be method-optimized for low-abundance cyclic nucleotides (matrix-dependent).
- Isomer Specificity: MRM workflows may distinguish 2'3' vs 3'3-cGAMP when standards and chromatography feasibility support it.
- QC Targets (Fit-For-Purpose): QC precision targets such as CV ≤10% and strong calibration linearity can be set based on matrix and study needs (IDMS optional).
- Stability Control: Immediate quenching with cold acidic extraction helps preserve labile cyclic nucleotides by minimizing PDE activity.
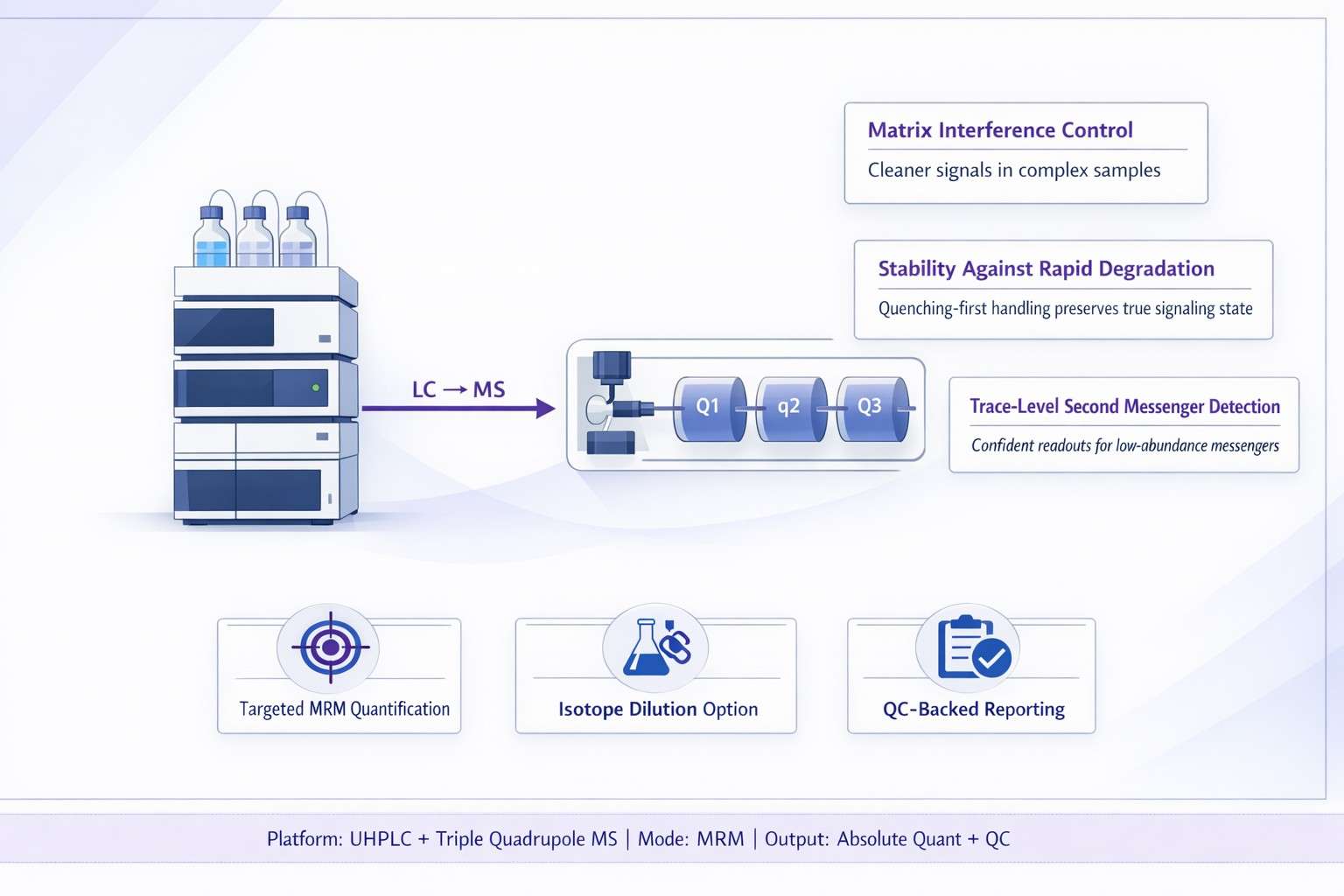
Our LC-MS/MS Workflow for Cyclic Nucleotide Analysis
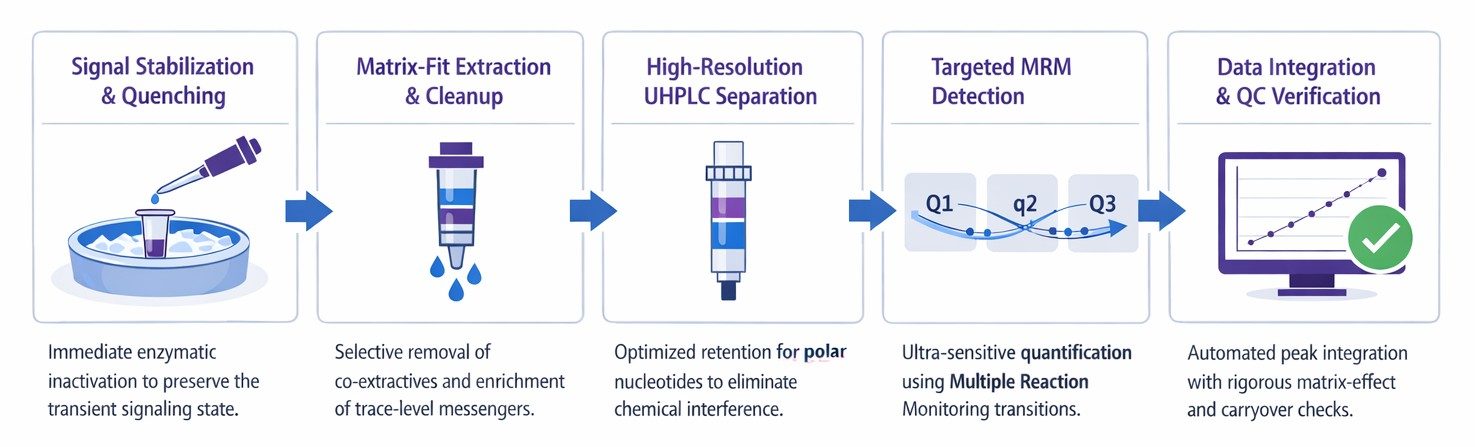
Analytical Platform and Key Parameters
We quantify cyclic nucleotides using a targeted UHPLC–triple quadrupole LC-MS/MS platform configured for MRM, designed for low-abundance second messengers in complex research matrices.
Core instruments
- Agilent 1290 UHPLC System
- Agilent 6495C Triple Quadrupole LC/MS (MRM)
Key parameters (typical approach)
| Parameter |
Typical Setting / Approach |
Why It Matters |
| Acquisition |
MRM |
High specificity in complex matrices |
| Ionization |
ESI (polarity optimized per analyte/matrix) |
Supports sensitivity and comparability |
| LC Strategy |
Polar-friendly UHPLC (e.g., HILIC or equivalent) |
Improves retention, reduces interferences |
| Quantification |
Calibration + QC; IDMS optional |
Stronger accuracy/precision |
| Matrix Controls |
Optional matrix-effect & spike/recovery checks |
Better cross-matrix confidence |
| cGAMP Specificity |
Optional isomer-aware support (feasibility-dependent) |
Distinguishes 2'3' vs 3'3 when needed |
Sample Submission and Preparation Guidelines
Use the table below as a practical starting point. If you have limited material, mixed matrices, or proprietary buffers, we can scope a matrix-fit plan.
| Sample Type |
Typical Suggested Amount (Starting Point) |
Container |
Storage & Shipping (General) |
Notes |
| Cell pellet |
1–5 million cells (quench immediately) |
Screw-cap tube |
Freeze; ship cold/frozen |
Minimize freeze–thaw |
| Cell lysate |
100–200 μL |
Low-bind tube |
Freeze; ship cold/frozen |
Provide buffer composition if possible |
| Tissue |
20–50 mg (flash-frozen) |
Cryovial |
Freeze; ship cold/frozen |
Record wet weight |
| Plasma / serum |
100–200 μL |
Screw-cap tube |
Freeze preferred |
Note anticoagulant (if applicable) |
| CSF |
50–100 μL |
Low-bind tube |
Freeze; ship cold/frozen |
Low abundance—enhanced QC recommended |
| Culture media / supernatant |
200–500 μL |
Screw-cap tube |
Freeze preferred |
Note medium/serum type |
Technical Pro-tip (Handling & Quenching): Cyclic nucleotides are sensitive to endogenous phosphodiesterases (PDEs). To preserve the true signaling state, quench immediately using cold acidic extraction (e.g., ~0.1 M HCl or HClO₄) and keep samples cold/frozen before shipping.
What You Receive
- Quantitative results tables (CSV/Excel): concentrations or response ratios, sample IDs, and normalization fields (as defined during scoping)
- Calibration summary: curve fit details and quant range notes (project-defined)
- QC documentation: QC performance, blanks/carryover checks, and matrix-control results when included
- Method snapshot: platform + workflow description suitable for internal documentation
- Optional raw outputs: chromatograms/integration exports (as requested)
Applications of cAMP, cGMP, and cGAMP Quantification
MS-CETSA functional proteomics uncovers new DNA-repair programs leading to Gemcitabine resistance
Nordlund, P., Liang, Y. Y., et al.
Journal: Research Square
Year: 2024
DOI: https://doi.org/10.21203/rs.3.rs-4820265/v1
Teriflunomide/leflunomide synergize with chemotherapeutics by decreasing mitochondrial fragmentation via DRP1 in SCLC
Mirzapoiazova, T., et al.
Journal: iScience
Year: 2024
DOI: https://doi.org/10.1016/j.isci.2024.110132
Unscheduled m6A Deposition in RNA via m6ATP Incorporation by DNA Polymerases
Qu, F., et al.
Journal: International Journal of Molecular Sciences
Year: 2025
DOI: https://doi.org/10.3390/ijms26199263
Hierarchical glycolytic pathways control the carbohydrate utilization regulator in human gut Bacteroides
Kabonick, S. G., et al.
Journal: Nature Communications
Year: 2025
DOI: https://doi.org/10.1038/s41467-025-59704-3